Abstract
Complete endoscopic evaluation of the small bowel is challenging due to its length and anatomy. Although several advances have been made to achieve diagnostic and therapeutic goals, including double-balloon enteroscopy, single-balloon enteroscopy, and spiral enteroscopy, video capsule endoscopy (VCE) remains the least invasive tool for complete visualization of the small bowel and is the preferred method for initial diagnostic evaluation. At present, interpretation of VCE data requires manual annotation of landmarks and abnormalities in recorded videos, which can be time consuming. Computer-assisted diagnostic systems using artificial intelligence may help to optimize VCE reading efficiency by reducing the need for manual annotation. Here we present a large VCE data set compiled from studies performed at two United States hospitals in Providence, Rhode Island, including 424 VCE studies and 5,247,588 total labeled images. In conjunction with existing published data sets, these files may aid in the development of algorithms to further improve VCE.
Measurement(s) | organ |
Technology Type(s) | Videocapsule Endoscopy |
Sample Characteristic - Organism | Homo sapiens |
Sample Characteristic - Location | State of Rhode Island |
Similar content being viewed by others
Background & Summary
The small intestine is about 6 m in length and is the longest organ in the gastrointestinal (GI) tract. It begins at the gastric pylorus and ends at the ileocecal valve, and is comprised of three segments known as the duodenum, jejunum, and ileum. The duodenum is about 25 cm in length and is divided into four sections known as the bulb, second or descending portion, third or transverse or horizontal portion, and fourth or ascending portion. The jejunum and ileum together measure about 5 m. Functionally, the small intestine is responsible for digestion and nutrient absorption. This is accomplished through a unique mucosal lining composed of finger-like projections, called villi, which augments the small intestine surface area by 600- to 1000-fold, totaling 250 to 400 m2 1. Disorders affecting the small intestine can be quite debilitating, often resulting in significant nutritional deficiencies and diarrheal illness, and can include Celiac disease, Crohn’s disease, malignancies, and other diseases of vascular and infectious etiologies. Gastrointestinal bleeding from the small intestine is infrequent, accounting for an estimated 3–5% of GI bleeds, and is often challenging to diagnose and treat2. Video capsule endoscopy (VCE) is used in the assessment of obscure GI bleeding and as a complementary diagnostic tool in other small bowel disorders when other endoscopic and imaging modalities are unrevealing.
VCE is usually performed via the use of a swallowed or endoscopically placed disposable plastic capsule containing a battery, camera, transmitter, and light source, which wirelessly transmits images via a sensor array belt worn by the patient to a data recorder; one system stores the images with the camera and downloads them later for review. Several VCE systems have been developed by manufacturers including Olympus America, Given Imaging, Intromedic Company, Ankon Technologies, Chongqing Science, and CapsoVision3,4,5. Technological improvements have allowed for wider fields of view (140°–360°), improved battery life, increased numbers of enclosed cameras, and variable frame rates (2–35 frames per second) that can be adjusted based on the speed of transit through the GI tract, capturing fewer images with slow transit and more images with faster transit5. Each system uses proprietary software to process images for review by the physician5. While handheld viewers allow for real-time review of images with some systems, VCE studies are most often reviewed after the capsule has passed completely through the small intestine. Ultimately, 8–12 hours of video and over 50,000–60,000 images may be generated in an individual study5. Image review is aided by software that allows for grouped viewing of 2–4 images at one time. Red pixel detection can also help to identify possible areas of bleeding. VCE image assessment remains time consuming. Review for the identification of anatomical landmarks and abnormalities requires up to 30–90 minutes per study. Fatigue related to the sustained and prolonged degree of concentration necessary to review such a large number of images may lead to errors, missed lesions, as well as inter- and intra-observer variation.
Computer-assisted diagnostic systems using artificial intelligence may help to address these challenges through automated data analysis, reducing VCE study assessment time, limiting required manual annotations, and improving both lesion detection and capsule localization3. Annotated VCE data sets may help to achieve this by aiding in the development of machine learning algorithms. There are currently few such VCE data sets, and most prioritize lesion detection3,6. Here we present the Rhode Island Gastroenterology VCE data set, a publicly available annotated data set, which contains 424 videos with 5,247,588 total labeled images denoting 4 anatomic classifications. To our knowledge, this is the largest such public data set currently available. While not every video in this data set was completely normal, annotations of abnormalities are not included here. Our focus was instead on landmark identification, a key initial step in the assessment of VCE studies which sets the foundation for correctly identifying and locating abnormalities7,8. Identifying abnormalities on VCE without adequate localization within the GI tract limits the ability to direct subsequent endoscopic interventions. This is particularly true in the small bowel where precise localization remains challenging and even minor improvements may be beneficial. Furthermore, machine learning guided identification of landmarks may help to limit variability among physicians with varying levels of expertise, thus leading to improved standardization9. Finally, identifying landmarks of segments within the GI tract at present requires a significant amount of manual annotation. Reducing the time required for this will improve the efficiency of VCE study interpretation10. Our hope is that this current data set will be most useful in improving landmark identification by artificial intelligence systems. Additional future endeavors to annotate abnormalities in this data set may help to improve artificial intelligence assisted lesion detection.
Methods
Data collection
The Rhode Island GI VCE data set was obtained from the VCE procedures performed at Rhode Island Hospital (RIH) and The Miriam Hospital (TMH) from January 2016 to December 2020. The VCE device used was the PillCamTM SB3 capsule, which was used together with its belt and recorder. Following complete image collection, images were downloaded for review and de-identified. Several patients underwent more than one VCE study, however, due to the de-identification process, these particular studies cannot be identified and were each treated as unique studies. This project was reviewed by the institutional review board of RIH and TMH (RIH IRB 1: IRB00000396, RIH IRB 2: IRB00004624, and TMH IRB: IRB00000482). As the project was granted a waiver of informed consent, these data were retrospectively obtained without the patients’ written consent. There is no protected health information contained in the data set and there are no restrictions on the use or disclosure of de-identified health information11. As such, these data may be shared under the Health Insurance Portability and Accountability Act Privacy Rule (HIPAA Privacy Rule).
Data de-identification
The RAPIDTM Reader or PillcamTM software is developed and supplied by the device manufacturer (Medtronic), and is used to open the proprietary video file downloaded from the PillCamTM system. To remove personal patient information from the video files, capsule studies were exported using the de-identify function in the RAPID reading software.
Data conversion
Following the de-identification process, the exported files remain in the proprietary video format. As a result, they must be converted into images (.png) using FFmpeg software (available from http://ffmpeg.org/). The original resolution is 320 × 320 pixels.
Data labeling
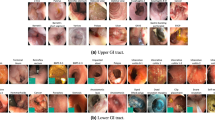
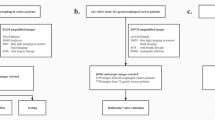
VCE images were classified into four classes by anatomical organ, including esophagus, stomach, small bowel, and colon. We identified the first image of each organ and the last image of each organ (the image before the first image of the next organ, except in the case of the colon). All images from the first image to the last image of each organ were labeled accordingly. The images that could not be classified into one of the four above described anatomical organs were removed. The majority of the removed images were before the first image of esophagus. To ensure the quality of the data set, the data were reviewed by two physicians specializing in gastroenterology. The first physician labeled the first images of each anatomical organ during the standard process of VCE interpretation. The second physician reviewed and validated the initial results. If there were any differences, a consensus between the two physicians was reached.
Inclusion and exclusion criteria
We only included VCE studies in which patients swallowed the capsule themselves. Endoscopy-deployed VCE studies were excluding from this data set. Furthermore, we only included VCE studies in which the images could be sequentially labeled into four anatomical organs.
Data Records
In the Rhode Island GI VCE data set, there are 424 VCE studies consisting of 5,247,588 images in total. Each study is saved in the form of a zip file containing four sub-folders for each anatomical organ (1 esophagus, 2 stomach, 3 small bowel, and 4 colon); each sub-folder represents a labeled anatomical organ in the gastrointestinal tract. The average number and total number of images per anatomical organ are presented in Fig. 1 and Table 1. The representative images for each anatomical organ shown in Fig. 2 were taken from the first sample (s001). These data are available at Figshare12.
Technical Validation
To provide a baseline performance for the classification model, we used the Inception ResNet V2 architecture to create a predictive model based on a down-sampling training set13. Inception Resnet V2 is among the most popular models for image classification that can provide very good performance and convergence speed, which are derived from the strengths of the inception model and residual networks. Based on the Pareto principle (80/20 rule), the data set of 424 samples was randomly separated into (1) training/validation set of 339 samples (80%) and 4,193,038 images (79.9%), and (2) testing set of 85 samples (20%) and 1,054,550 images (20.1%). In the training/validation set, the smallest class is esophagus with 9,061 images (0.2%). Since the minority class is less than 1% of the total number of images, the degree of class imbalance is considered extreme. As a result, we performed down-sampling on other classes to reduce class imbalance issues such that the number of images of the majority classes (stomach, small bowel, and colon) was reduced to 11,508, 32,252, and 11,707 images, respectively. To reduce the degree of imbalance, the down-sampling was done by randomly selecting images (1 out of 400 for small bowel and 1 out of 100 for the two classes) per class in each sample. It is worth noting that the down-sampling provide an additional benefit in reducing the training data size and computational resources for training the deep learning model. Then, we randomly split the training/validation set into a training set (75%) and a validation set (25%); the splitting proportion was set to optimize the training time within a reasonable time period. The number of images per data set is summarized in Table 2. The Inception ResNet V2 model was used to train the model; the trained model was then used to predict the anatomical organs on the full testing set.
The testing performance is presented in Table 3. The overall accuracy is 97.1%, however, the classification performance is highly class dependent. The small bowel, which is often the primary organ under investigation in VCE studies, demonstrates the best performance (precision = 0.9939, recall = 0.9793, and F1 score = 0.9865). Mis-classification (for example, predicting stomach from an image of the colon) may lead to complex problems in practice when attempting to identify the first image of each anatomical organ. The confusion matrix (Fig. 3) shows that the mis-classification rate is relatively low.
Usage Notes
The data set is being shared under the Creative Commons Attribution 4.0 International Public license, which allows users to use, share, adapt, distribute, and reproduce the data set with the appropriate credit given to the original authors and the data source. More details regarding the license can be found in https://creativecommons.org.
The image names in each of the VCE studies are in numerical order. We did not exclude any images from the esophagus to the colon. As a results, some images contain abnormalities such as bleeding, erosions, polypoid lesions, and other GI lesions.
Code availability
The code and information needed for training and testing the deep learning model presented in the technical validation section are published on GitHub, which can be accessed via https://github.com/acharoen/Rhode-Island-GI-VCE-Technical-Validation. The pre-trained model used to generate the predictions (InceptionResNetV2.h5) is stored in the same data repository12.
References
Campbell, J., Berry, J. & Liang, Y. Chapter 71 - anatomy and physiology of the small intestine. In Yeo, C. J. (ed.) Shackelford’s Surgery of the Alimentary Tract, 2 Volume Set (Eighth Edition), 817–841, https://doi.org/10.1016/B978-0-323-40232-3.00071-6 (Elsevier, Philadelphia, 2019).
Lewis, B. S. Small intestinal bleeding. Gastroenterol. Clin. North Am. 29, 67–95, https://doi.org/10.1016/S0889-8553(05)70108-4 (2000).
Smedsrud, P. H. et al. Kvasir-capsule, a video capsule endoscopy dataset. Sci. Data 8, https://doi.org/10.1038/s41597-021-00920-z (2021).
Wang, A. et al. Wireless capsule endoscopy. Gastrointest. Endosc. 78, 805–815, https://doi.org/10.1016/j.gie.2013.06.026 (2013).
Akpunonu, B., Hummell, J., Akpunonu, J. D. & Ud Din, S. Capsule endoscopy in gastrointestinal disease: Evaluation, diagnosis, and treatment. Clevel. Clin. J. Medicine 89, 200–211, https://doi.org/10.3949/ccjm.89a.20061. https://www.ccjm.org/content/89/4/200.full.pdf (2022).
Koulaouzidis, A. et al. KID project: an internet-based digital video atlas of capsule endoscopy for research purposes. Endosc. Int. Open 05, E477–E483, https://doi.org/10.1055/s-0043-105488 (2017).
Mitselos, I. V. & Christodoulou, D. K. What defines quality in small bowel capsule endoscopy. Annals Transl. Medicine 6 (2018).
Koulaouzidis, A., Dabos, K., Philipper, M., Toth, E. & Keuchel, M. How should we do colon capsule endoscopy reading: a practical guide. Ther. Adv. Gastrointest. Endosc. 14, 26317745211001983, https://doi.org/10.1177/26317745211001983, PMID:33817637 (2021).
Schelde-Olesen, B. et al. Inter-observer agreement in landmark and flexure identification in colon capsule endoscopy. European Society of COLOPROCTOLOGY, ESCP; Conference date: 21-09-2022 Through 23-09-2022 (2022).
Ayyoubi Nezhad, S., Khatibi, T. & Sohrabi, M. Proposing novel data analytics method for anatomical landmark identification from endoscopic video frames. J. healthcare engineering 2022 (2022).
U.S. Department of Health & Human Services. Summary of the hipaa privacy rule, https://www.hhs.gov/hipaa/for-professionals/privacy/laws-regulations/index.html#:~:text=The%20Privacy%20Rule%20protects%20all,health%20information%20(PHI). Last accessed 22 June 2022 (2013).
Charoen, A. Rhode island gastroenterology video capsule endoscopy data set, figshare https://doi.org/10.6084/m9.figshare.c.6071216.v1 (2022).
Szegedy, C., Ioffe, S. & Vanhoucke, V. Inception-v4, inception-resnet and the impact of residual connections on learning. CoRR abs/1602.07261, https://doi.org/10.48550/arXiv.1602.07261 (2016).
Acknowledgements
This material is based upon work supported by the Google Cloud Research Credits program with award GCP19980904, and Brown Physicians, Inc.
Author information
Authors and Affiliations
Contributions
A.C. and H.G.R. initiated the project, A.C., P.F. and H.G.R. labeled the data. A.C., S.T., N.W. conducted the experiments and analyzed the results. A.C. and A.G. drafted the initial manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Charoen, A., Guo, A., Fangsaard, P. et al. Rhode Island gastroenterology video capsule endoscopy data set. Sci Data 9, 602 (2022). https://doi.org/10.1038/s41597-022-01726-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-022-01726-3
This article is cited by
-
Public Imaging Datasets of Gastrointestinal Endoscopy for Artificial Intelligence: a Review
Journal of Digital Imaging (2023)
-
Utilisation of a wrinkled film-based structure for the sensing measurement of a vibro-impact capsule robot
Meccanica (2023)