Abstract
Lignin is one of the most abundant biopolymers in nature and has great potential to be transformed into high-value chemicals. However, the limited availability of molecular structure data hinders its potential industrial applications. Herein, we present the Lignin Structural (LGS) Dataset that includes the molecular structure of milled wood lignin focusing on two major monomeric units (coniferyl and syringyl), and the six most common interunit linkages (phenylpropane β-aryl ether, resinol, phenylcoumaran, biphenyl, dibenzodioxocin, and diaryl ether). The dataset constitutes a unique resource that covers a part of lignin’s chemical space characterized by polymer chains with lengths in the range of 3 to 25 monomer units. Structural data were generated using a sequence-controlled polymer generation approach that was calibrated to match experimental lignin properties. The LGS dataset includes 60 K newly generated lignin structures that match with high accuracy (~90%) the experimentally determined structural compositions available in the literature. The LGS dataset is a valuable resource to advance lignin chemistry research, including computational simulation approaches and predictive modelling.
Measurement(s) | molecular structure |
Technology Type(s) | Computer Modeling |
Factor Type(s) | monomer ratio • bond frequency • degree of polymerization |
Sample Characteristic - Organism | coniferous (softwood) • deciduous (hardwood) |
Similar content being viewed by others
Background & Summary
Lignin is one of the most abundant biopolymers in nature followed by cellulose. Natural biopolymers such as DNA contain well-defined arrangements of monomers and linkages that can be consistently determined through experimental characterization. In contrast, lignin’s monomer sequence and structure are not well-defined and present extremely complex chemical structures resulting from an irregular biosynthesis process1,2. Lignin is not used to its fullest potential due to uncertainties in the characterization of its primary molecular structure.
Lignin structures are formed primarily by three main repeating units (monolignols), arranged in random sequences. The major contributors to lignin’s primary structure are p-coumaryl (H), coniferyl (G), and sinapyl (S) alcohols3,4. The monomers radically couple and cross-link combinatorially5 to form multifaceted lignin structures. During lignin biosynthesis (or lignification), monomers are transported to the cell wall where they are polymerized in a combinatorial fashion by free-radical coupling mechanisms in a reaction mediated by peroxidases, generating a variety of structures within the lignin polymer3,6,7. Lignin’s structural composition varies between plant species and changes depending on the tissue, cell location or environmental conditions8,9,10. In addition, major structural changes occur during the lignin separation process11,12. During the last decades, various applications of lignin have been investigated, either as a macromolecule or after depolymerization into lower molecular weight compounds. Although there are numerous studies on lignin polymerization process and depolymerization13, there is still a lack of curated lignin structure datasets that can support lignin chemistry research.
Molecular structure and the length of monomer chains determine polymer properties (e.g., strength, solubility, heat resistance). Since there may be multiple molecular structures consistent with a given average chemical composition, elucidating the correct molecular structure and structural variations responsible of specific property profiles is a key challenge in lignin chemistry. Current advances in polymer science and computational modelling14 provide new insight on the structural features of this macromolecule and guide bioengineering strategies for achieving structures with targeted properties. Experimental techniques such as spectroscopy (e.g., NMR and FT-IR) or wet chemistry (e.g., thioacidolysis, nitrobenzene oxidation, and hydrogenolysis) are key to reveal the molecular features of lignin1,15,16 and provide statistical descriptions of the polymer structure. Alternatively, computational methods can be used to predict lignin structure and properties. Multiple research efforts have modelled the lignification process using a variety of simulation techniques parameterized by experimental data on monomer and bond distributions17,18,19,20,21. Simulation studies on lignin structure formation are often carried out by adding monomeric units to reactive sites in the polymer17,18,19,22.
This paper describes a novel lignin polymer dataset, herein referred as lignin structural dataset (LGS), containing computer-generated molecular structures of milled wood lignin (MWL). The LGS dataset includes structural isomers for experimental data on spruce (softwood) and birch (hardwood) MWL. This resource provides data for 60 K lignin molecules (6.3 K softwood structures and 53.7 K hardwood structures) with varying degree of polymerization (DP) in the range of 3 to 25. The LGS dataset, available on Figshare23, can be readily used in studies aimed at understanding lignin’s structural properties using parameterized force fields24 and molecular dynamics approaches21,25. The structural information contained in the dataset provides a unique resource for simulating the depolymerization of lignin into lower molecular weight compounds. In addition to the structure data, the paper also describes a new computational approach to sequence-controlled polymer generation. Sequence-controlled polymers are macromolecules in which monomer units of different chemical nature are arranged in an orderly fashion26. A polymer formed by radical chain‐growth polymerization is also considered as a sequence‐controlled polymer, although it is also a nonuniform polymer with chains of different lengths and slightly different composition27. The tool can generate chemically correct and legible 2D structures of the MWL for hardwood, softwood and herbaceous wood types. The code used to generate and validate lignin structures is also provided for reproducibility.
Methods
The computational approach used to generate the LGS dataset is based on sequence-controlled polymer growth parameterized to match experimental lignin structure data. Lignin’s structural heterogeneity can be characterized by the inter-unit linkages and functional groups attached to phenyl propane units. Inter-unit linkages are present in two major categories corresponding to ether bond linkages (e.g., β-O-4, α- O-4, 4-O-5) and carbon-carbon linkages (e.g., β–β, β-5, 5–5). The relative abundance of each linkage in native lignin varies from plant to plant with the β-O-4 linkage being the most abundant28,29. The major functional groups present in lignin structures include hydroxyl, methoxy, carbonyl and carboxylic groups. The proportion of these groups in phenyl propane units depends on the species genetic origin and isolation processes.
Lignin structure compositions were analyzed from experimental studies on MWL of different wood species. Table 1 summarizes experimentally determined frequencies of primary monolignols and linkages in softwood and hardwood species.
Experimental characterization3,5,10,30,31,32 distinguishes two main classes of lignin biopolymers, G type lignin which is typical of conifers or softwood, and SG type lignin which is formed in deciduous plants or hardwood. The presence of H monomers is minimal in both of these lignin types33. The main structural variations of lignin include straight chain (linear) structures, branching structures, and cross linking with carbohydrates34. Two forms of coupling reactions could occur during lignin biosynthesis. First, linear reactions which contribute to extending the macromolecule by coupling new monomer units with β-O-4, β–β and β-5 linkages to form monomer-monomer and oligomer-monomer couplings. Second, branching reactions that form oligomer-oligomer couplings with 5-5 and 4-O-5 linkages3,26,30,35. It is important to note that units with 5-5 linkage have an alternate form with an 8-member ring structure named dibenzodioxocin (DBDO)36,37 which is predominantly present in softwood lignin. Spirodienone (β-1′/α-O-α′) has also been reported as a possible linkage with low prevalence (~3–4 in 100 phenylpropanoid units)3,38.
Generation of lignin structures
The structure generation approach used to create the LGS dataset doesn’t assume that lignin conforms to a specific sequence of monomers and bond types. Instead, our work assumes that lignin structures are created as the result of a random or near-random coupling of primary monomers, where each monomer can couple in any of all possible ways. To capture lignin’s structural variability, we have developed a generative model based on a combinatorial approach that uses a sequence-controlled method to create theoretical lignin structures. The algorithm starts by defining the primary monomer units followed by the application of molecular connectivity rules based on experimental studies3,5,10,30 to generate plausible monomer sequences. Subsequent steps generate all possible molecular arrangements with similar structural features (i.e., monomer ratio and linkages). The validity of the resulting lignin polymers is then corroborated by comparing their features (e.g., functional groups, linkages, and end groups) with experimental data as well as by matching with molecular fingerprints of structures extracted from recent studies20,21 on lignin structure simulation. Figure 1 summarizes the workflow used for structure generation with details on the required input information and the structure of the generated LGS dataset.
The input information needed to execute the workflow includes the S/G ratio, bond ratio, and degree of polymerization. Experimental characterization data used to bootstrap the structure generation were obtained from analytical results reported in previous studies5,30,39,40 on softwood and hardwood lignin samples. The degree of polymerization (DP) is computed as the ratio between the weight average molecular weight (Mw) of lignin polymer and the weight average molecular weight of phenylpropane monomer (M0) (i.e., DP = Mw/M0)41. The degree of polymerization represents the number of monomer units in each lignin structure. The structure generation workflow starts with sequencing lignin monomers and the generation of linkages, followed by the creation of topological matrices and molecular graph representations. In subsequent steps, graphs are converted to canonical molecular structures and the consistency of the generated structures is verified by comparison with experimental observations and simulated data. Finally, all validated structures are assembled to create the LGS dataset. The next subsections provide specific details on each workflow step.
Sequencing lignin monomers and conditional linkage generation
Phenyl propane units (Fig. 2) form the core backbone of the lignin structure42. The base unit in the backbone contains structural features of p-coumaryl alcohol (H), in which an allylic alcohol moiety is connected to the para position of phenolic ring. The three carbon atoms present in the aliphatic side chain are located at α, β and γ positions respectively. During lignin structure generation, the H monomer is considered as the base unit whereas coniferyl alcohol (G monomer) and syringyl alcohol (S monomer) are modifications of the base unit with additions of one (position 3) and two (position 3 and 5) methoxy groups, respectively.
The lignin structure generator uses a permutation approach to create lignin structures including all possible combinations of H, G, and S monomers. However, since most available experimental data corresponds to G and SG type lignin structures, only S and G monomers were considered for inclusion in the LGS dataset. Table 2 summarizes the linkage patterns used to generate lignin structures. The sequence generation process creates all possible arrangements of the primary monomer units to produce a lignin chain. For instance, in a G type structure with five monomers, the repeating unit will be [G, G, G, G, G] with a single monomer sequence whose length is controlled by DP. For SG type structures, a set of permutations based on the S/G ratio and DP is used to generate all possible monomer arrangements.
The sequence generation code implements Heap’s permutation algorithm43 using a recursive function adjusted to generate all possible permutations of n monomer units (Fig. 3). Although there are nPr possible monomer permutations, filtering duplicate arrangements reduces the number of candidate structures. For instance, running the sequence generation algorithm with 5 monomer units generates 10 unique sequences for a S/G ratio of 1.8 as illustrated in Fig. 3.
The linkage sequence is generated using a similar approach, except that the bonding position is rearranged based on monomer labels (i.e., G, S) and possible linkages between monomers. Table 3 presents the coupling patterns for monomers and oligomers reported in the literature3,5,30,39. Structures are linearly coupled when one of the favored sites is the β position. Coupling reactions between oligomers produce 5-5 and 4-O-5 branched structures.
Experimental analysis data from previous studies5,30,44,45 have shown different branching structure patterns associated with 5-5 and 4-O-5 linkages in native/MWL lignin. The different branching patterns considered in this model are phenolic 5-545, phenolic 4-O-55,30,45, phenolic dibenzodioxocin (DBDO)5,45, non-phenolic DBDO30,44,45 and etherified 5-545.
Generation of topological matrices and molecular graph representations
The structure generation mimics the monomer sequencing in chain-growth polymerization by assigning probabilities to the presence of specific monomer pairs. Resulting molecular structures can be represented as graphs where nodes represent monomer units, and the edges correspond to covalent bonds formed during the polymerization process. For example, in case of outgoing edges for linkage type β-O-4, the β position of the monomer represented in the first node is connected to the O-atom attached to the 4th carbon in the ring structure of the monomer in the second node. Studies on lignin structure report that monomers are bidirectionally linked5. To account for link directionality, labels are stored as tuples of variables (e.g., (G, S) (S, G) (G, S) (S, G)) that represent parent-child relationship with bonds represented as directed edges denoting the linkage direction. Monomer sequences and bond patterns generated in the previous step are processed to create linear and branched structures. Linear chains are created by adding monomers one-by-one to the polymer (i.e., endwise lignin growth). Branching chains are formed by coupling fragmented linear chains (i.e., two lignin oligomers) using the coupling patterns described in Table 3. Figures 4 and 5 illustrate the development of linear and branched structures.
Molecular graphs can be efficiently represented in tabular form as a pair of topological matrices46 that define the relationship between monomer units. The information in the topological matrices includes linkages (i.e., adjacency matrix) and bond types (i.e., connectivity matrix). These matrices identify the bond direction and occupied bond position for each graph node (e.g., β carbon as ‘B’, 5th carbon for G unit as ‘5’, 4-O position in the ring as ‘4’). Figure 6 shows the directed graph and 2D molecular representations of a lignin polymer together with the structure encoding using topological matrices. Additional information describing the format of the topological matrices can be found in the Supporting Information (Figure F).
Generation of molecular structures and consistency validation
The molecular structure of a lignin is represented using the canonical SMILES (Simplified Molecular Input Line Entry System) notation which represents the molecular structure in a string format47. Directed graphs are converted to canonical SMILES by generating the monomer molecular graph and mapping the respective linkages using the Chemistry Development Toolkit (CDK)48, a widely used open-source cheminformatics toolkit. Details on the software architecture used to implement the LGS generation tool are provided in the supporting Information (Figures A and B). The LGS tool allows the user to generate different structural variations for a given set of experimental observations by configuring the required parameters such as monomer ratio (S, G and H), bond frequencies (β-O-4, β–β, β-5, 4-O-5, 5-5 and DBDO) in the project configuration file (project-config.yaml). Finally, generated structures are checked for the acceptance criteria that include structures with unique and valid SMILES representation; and number of nodes matching the requested degree of polymerization. A complete dataset description and structure validations from experimental results are provided in the following sections.
Data Records
The LGS dataset provides structural information for 60 K unique lignin polymers including G and SG type structures with degree of polymerization in the range of 3 to 25. The LGS dataset can be accessed from Figshare23. The subset of spruce MWL structures (G type) contains 6.3 K polymers whereas the remaining 53.7 K polymers correspond to birch MWL structures (SG type). Evaluated features of softwood and hardwood MWL structures from the dataset align with structural features reported in experimental studies on lignin structural composition (Table 1). Figure 7 shows the linkage counts for the generated structures. The β-O-4 linkage is predominant across both types of wood with percentages in the range of 50-65%, which agrees with the experimental data summarized in Table 1. For G type structures, the β5/55/DBDO linkages are predominant (up to 6%) whereas 4-O-5 linkages are more frequent in SG type structures (1-6%).
Figure 8 summarizes the main structural features of the macromolecules included in the LGS dataset. Figure 8(a) depicts the ratio of branched structures present in relation to DP in the dataset. Linear chains are the predominant structures for DP up to 4 for both G type and SG type structures. In G type structures, the degree of branching increases abruptly up to 85% when DP is greater than 10. In contrast, for SG structures increasing DP results in branching degrees in the range of 15–30%. The abrupt transition observed for G type structures is due to the presence of vacant 5th carbon in the ring which enables the possibility of forming 5-5 or 4-O-5 structures with increased number of monomer units.
Structural features in the LGS dataset (a) Percentage of branching structures, (b) Functional group counts for structures with a degree of polymerization of 20, (c,d) Clustered heatmap of the correlation of bond frequencies and DP in the data subsets corresponding to G and SG type lignin, respectively.
The analysis of functional group counts reveals further structural variations across G and SG type structures. Figure 8(b) depicts the variation in functional group counts for G and SG structures with a degree of polymerization of 20. Since β-O-4 is the major linkage present in both structure types, phenolic hydroxyl groups are mostly condensed. The higher frequency of resinol (β-β/γ-O-α/α-O-γ) structures reduces the number of free primary aliphatic hydroxyl groups in SG type structures. The presence of methoxy groups in SG structures is higher than for G structures. These observations align with previous studies on lignin’s structural charateristics12.
Figure 8(c,d) visualize the association of structural features (i.e., linkage and DP) across G and SG type structures. The analysis of the clustered heatmaps reveals structural commonalities and variation patterns. The β-O-4 linkage is strongly correlated with DP in both structure types. In G type structures, β-β bonds have higher correlation with 5-5 and DBDO linkages. The number of β-β bonds restricts the chain growth linearly and generates shorter oligomers which can then couple by 5-5 and 4-O-5 linkages to form branched structures. For SG type structures, the 4-O-5 linkage has higher correlation with DP relative to G structures. This is due to the presence of a methoxy group in the ring structure of syringyl alcohol that limits the formation of 5-5 or DBDO structures in favor of 4-O-5 structures.
File format
The LGS dataset provides lignin structures in three different formats to facilitate the compatibility with a variety of computational chemistry software platforms (e.g., ADF49, Avogadro50) and data analytics tools:
-
Application specific files: Molecular data files (MOL), which is a widely used chemical structure file format supported by most software packages for molecular dynamics. The data file contains 3D conformation of the generated molecule, which can be used in molecular editors such as Avogadro to visualize and analyze the spatial arrangements.
-
Tabular files: Text files using the Comma Separated Value (CSV) format that represent each molecular graph using connectivity and adjacency matrices.
-
Key/value file: Text files using the JavaScript Object Notation (JSON51) format that provide comprehensive information that includes the definition of molecular structures using SMILES together with properties such as molecular weight, functional group counts and bond ratios. This file also serves as catalog of the structural information with respect to specific DP such as S/G ratio, bond frequencies, SMILES string, etc., Lignin id (lg_id) in the JSON object provides a unique identifier to locate the properties of specific structures in *.mol and *.csv files, respectively.
Detailed file structure definitions and examples for each of the three data formats are provided in the Supporting Information (Figures E, F and G).
Visualization of the LGS dataset
Figure 9 depicts the Tree MAP (TMAP) projection of calculated MinHash fingerprints (MHFP6)52 for a subset of G type lignin structures. The algorithm is an interactive data visualization method that maps high-dimensional chemistry data onto lower dimensional feature spaces. Each point in the visualization represents a single lignin structure and the color mapping denotes specific polymer properties (e.g., molecular weight, monomer count, linkages). The TMAP projection algorithm is combined with JSmol53 to visualize the lignin molecules in 2D and 3D representations together with their associated property data. A TMAP visualization for SG type structures is provided in the Supporting Information (Figure D). An interactive implementation of the TMAP visualization algorithm is available at https://labs.wsu.edu/pmrg/resources/lgsdataset.
Technical Validation
A detailed comparative analysis was performed to ensure that the properties of the generated lignin structures are consistent with experimental values reported in the literature. To this end, the structure of the generated polymers (e.g., bond compositions, functional groups, and end groups) were compared with properties derived from experimental observations for MWL from Spruce and Birch wood. Results in Table 4 corroborate the agreement between generated and experimental data with degrees of polymerization of 18 and 20. Overall, the molecular weight and interunit linkage ranges for generated structures are consistent with experimental values reported in previous studies7,54 with the main differences due to functional group counts.
Additional validation was performed by comparing the structures in the LGS dataset with structures generated using the Lignin-KMC model20. The Lignin-KMC model implements a structure generation approach based on kinetically controlled radical coupling reactions55. Figure 10 provides examples of lignin structures generated by each model for the same S/G ratio and degree of polymerization. Although the two lignin polymers are different, both KMC and LGS generated structures have similar features (e.g., linkage forms that includes β-aryl ether, resinol and phenyl coumaran).
The cheminformatics software package Datawarrior56 was used to inspect the differences across lignin structures with DP = 6 generated using each modeling approach. Molecular structures were encoded using binary fingerprints57 and projected onto a 2D space using t-distributed stochastic neighbor embedding (t-SNE)58. The three clusters identified in Fig. 11 group molecules with similar structural patterns. Structures in the LGS dataset overlap in each cluster with structures generated by Lignin-KMC, with small variations in the number of oligomers and cinnamyl alcohol end groups.
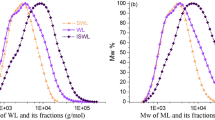
At a higher degree of polymerization (i.e., DP = 18), the LGS model generates molecular structures with higher frequency of β-O-4 linkages that result in molecular structures with longer oligomer chains relative to those generated by the lignin-KMC model. Unlike the KMC model, the LGS approach can generate dibenzodioxocin(5-5/β-O-4/α-O-4) linkages resulting in a more diverse set of lignin structures. Figure 12 shows the difference in the molecular weight and number of oligomers for the largest lignin chain generated by each model. The LGS approach generates structures that are in good agreement with the experimental observations for MWL with larger oligomers and higher β-O-4 frequencies relative to the lignin-KMC model.
Usage Notes
The LGS dataset constitutes a unique resource that provides primary structural data for milled wood lignin. The information included in the dataset can be used for molecular mechanics and predictive modelling studies on lignin. The dataset can also be used as a foundation to simulate lignin depolymerization into low molecular weight compounds. For instance, lignin depolymerization using deep eutectic solvents (DES) produces lower molecular weight compounds through a selective ether bond cleavage59 that can be simulated using the MWL structures included in the LGS dataset.
Along with MWL dataset, this study developed a computational framework to derive the structure of the lignin macromolecule by simulating longer polymer structures based on the molecular linkage patterns present on experimental data. Although spatial (i.e., 3D) arrangements of lignin macromolecules were not explored as part of this study, the current work provides a foundation for future studies and model extensions to incorporate these effects leading to a more comprehensive understanding of the molecular features of lignin.
Code availability
The LGS dataset and the lignin structure generator tool are available on Figshare23 and GitHub (https://github.com/sudhacheran/lignin-structure-generator) respectively. Moving forward, the database and accompanying tools will be periodically updated and extended. The latest version of the LGS dataset can be downloaded from: https://labs.wsu.edu/pmrg/resources/lgsdataset/.
References
Lu, Y. et al. Structural Characterization of Lignin and Its Degradation Products with Spectroscopic Methods. J Spectrosc 2017, 8951658, https://doi.org/10.1155/2017/8951658 (2017).
Gellerstedt, G. & Henriksson, G. In Monomers, Polymers and Composites from Renewable Resources (eds M. N., Belgacem & A., Gandini) 201–224 (Elsevier, 2008).
Ralph, J. et al. Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenyl- propanoids. Phytochemistry Reviews 3, 29–60, https://doi.org/10.1023/B:PHYT.0000047809.65444.a4 (2004).
Katahira, R., Elder, T. J. & Beckham, G. T. In Lignin Valorization: Emerging Approaches 1–20 (The Royal Society of Chemistry, 2018).
Ralph, J., Lapierre, C. & Boerjan, W. Lignin structure and its engineering. Curr Opin Biotech 56, 240–249, https://doi.org/10.1016/j.copbio.2019.02.019 (2019).
Boerjan, W., Ralph, J. & Baucher, M. Lignin biosynthesis. Annu Rev Plant Biol 54, 519–546, https://doi.org/10.1146/annurev.arplant.54.031902.134938 (2003).
Vanholme, R., Demedts, B., Morreel, K., Ralph, J. & Boerjan, W. Lignin biosynthesis and structure. Plant Physiol 153, 895–905, https://doi.org/10.1104/pp.110.155119 (2010).
Lourenco, A. et al. Lignin Composition and Structure Differs between Xylem, Phloem and Phellem in Quercus suber L. Front Plant Sci 7, ARTN 161210.3389/fpls.2016.01612 (2016).
Annunziata, M. G. What Is Lignin Made of? New Components Discovered. Plant Physiology 180, 1255–1255, https://doi.org/10.1104/pp.19.00561 (2019).
Nishimura, H., Kamiya, A., Nagata, T., Katahira, M. & Watanabe, T. Direct evidence for α ether linkage between lignin and carbohydrates in wood cell walls. Scientific Reports 8, 6538, https://doi.org/10.1038/s41598-018-24328-9 (2018).
Wen, J.-L., Wang, H.-M., Ma, C.-Y., Yuan, T.-Q. & Sun, R.-C. In 3 - Value-added products from lignin: IsolationValue-added products from lignin: Isolation, characterization and applications - Biomass, Biofuels, Biochemicals (eds T., Bhaskar & A., Pandey) 33–55 (Elsevier, 2021).
Huang, J., Fu, S. & Gan, L. In Chapter 2 - Structure and Characteristics of Lignin, Lignin Chemistry and Applications 25–50 (Elsevier, 2019).
Fache, M., Boutevin, B. & Caillol, S. Vanillin Production from Lignin and Its Use as a Renewable Chemical. Acs Sustainable Chemistry & Engineering 4, 35–46, https://doi.org/10.1021/acssuschemeng.5b01344 (2016).
Gartner, T. E. & Jayaraman, A. Modeling and Simulations of Polymers: A Roadmap. Macromolecules 52, 755–786, https://doi.org/10.1021/acs.macromol.8b01836 (2019).
Stark, N., Yelle, D. & Agarwal, U. In Techniques for Characterizing Lignin 49–66 (2015).
Agarwal, U. P., McSweeny, J. D. & Ralph, S. A. FT-Raman Investigation of Milled-Wood Lignins: Softwood, Hardwood, and Chemically Modified Black Spruce Lignins. J Wood Chem Technol 31, 324–344, https://doi.org/10.1080/02773813.2011.562338 (2011).
Dellon, L. D., Yanez, A. J., Li, W. J., Mabon, R. & Broadbelt, L. J. Computational Generation of Lignin Libraries from Diverse Biomass Sources. Energ Fuel 31, 8263–8274, https://doi.org/10.1021/acs.energyfuels.7b01150 (2017).
Yanez, A. J., Li, W. J., Mabon, R. & Broadbelt, L. J. A Stochastic Method to Generate Libraries of Structural Representations of Lignin. Energ Fuel 30, 5835–5845, https://doi.org/10.1021/acs.energyfuels.6b00966 (2016).
Glasser, W. G. & Glasser, H. R. Simulation of Reactions with Lignin by Computer (Simrel) .1. Polymerization of Coniferyl Alcohol Monomers. Macromolecules 7, 17–27, https://doi.org/10.1021/ma60037a005 (1974).
Orella, M. J. et al. Lignin-KMC: A Toolkit for Simulating Lignin Biosynthesis. Acs Sustainable Chemistry & Engineering 7, 18313–18322, https://doi.org/10.1021/acssuschemeng.9b03534 (2019).
Vermaas, J. V., Dellon, L. D., Broadbelt, L. J., Beckham, G. T. & Crowley, M. F. Automated Transformation of Lignin Topologies into Atomic Structures with LigninBuilder. ACS Sustainable Chemistry & Engineering 7, 3443–3453, https://doi.org/10.1021/acssuschemeng.8b05665 (2019).
Glasser, W. G. & Glasser, H. R. Simulation of Reactions with Lignin by Computer (Simrel) .2. Model for Softwood Lignin. Holzforschung 28, 5–11, https://doi.org/10.1515/hfsg.1974.28.1.5 (1974).
Eswaran, S. C. D., Subramaniam, S., Sanyal, U., Rallo, R. & Zhang, X. Milled wood Lignin Structural (LGS) Dataset. figshare https://doi.org/10.6084/m9.figshare.16915672.v2 (2022).
Vermaas, J. V., Petridis, L., Ralph, J., Crowley, M. F. & Beckham, G. T. Systematic parameterization of lignin for the CHARMM force field. Green Chem 21, 109–122, https://doi.org/10.1039/c8gc03209b (2019).
Zhang, L. & LeBoeuf, E. J. A molecular dynamics study of natural organic matter: 1. Lignin, kerogen and soot. Organic Geochemistry 40, 1132–1142, https://doi.org/10.1016/j.orggeochem.2009.08.002 (2009).
Achyuthan, K. E. et al. Supramolecular self-assembled chaos: polyphenolic lignin’s barrier to cost-effective lignocellulosic biofuels. Molecules 15, 8641–8688, https://doi.org/10.3390/molecules15118641 (2010).
Lutz, J. F. Defining the Field of Sequence-Controlled Polymers. Macromol Rapid Commun 38, https://doi.org/10.1002/marc.201700582 (2017).
Ma, R., Zhang, X., Wang, Y. & Zhang, X. New Insights Toward Quantitative Relationships between Lignin Reactivity to Monomers and Their Structural Characteristics. Chemsuschem 11, 2146–2155, https://doi.org/10.1002/cssc.201800550 (2018).
Yamaguchi, A., Mimura, N., Shirai, M. & Sato, O. Bond cleavage of lignin model compounds into aromatic monomers using supported metal catalysts in supercritical water. Scientific Reports 7, 46172, https://doi.org/10.1038/srep46172 (2017).
Balakshin, M. et al. Spruce milled wood lignin: linear, branched or cross-linked. Green Chem 22, 3985–4001, https://doi.org/10.1039/d0gc00926a (2020).
Holtzapple, M. T. In Encyclopedia of Food Sciences and Nutrition (Second Edition) (ed Benjamin Caballero) 3535–3542 (Academic Press, 2003).
Novaes, E., Kirst, M., Chiang, V., Winter-Sederoff, H. & Sederoff, R. Lignin and biomass: a negative correlation for wood formation and lignin content in trees. Plant Physiol 154, 555–561, https://doi.org/10.1104/pp.110.161281 (2010).
Zahed Mahmood, M. Y., M Jahangeer, M Riaz, A Ghaffar, I Javid. Lignin as Natural Antioxidant Capacity, Lignin - Trends and Applications. Matheus Poletto, IntechOpen, https://doi.org/10.5772/intechopen.73284 (March 12th 2018).
Tarasov, D., Leitch, M. & Fatehi, P. Lignin-carbohydrate complexes: properties, applications, analyses, and methods of extraction: a review. Biotechnol Biofuels 11, 269, https://doi.org/10.1186/s13068-018-1262-1 (2018).
Ana Lourenço, H. P. Compositional Variability of Lignin in Biomass. Lignin - Trends and Applications, pp. 65–98, https://doi.org/10.5772/intechopen.71208 (2018).
Karhunen, P., Rummakko, P., Sipila, J., Brunow, G. & Kilpelainen, I. Dibenzodioxocins - a Novel Type of Linkage in Softwood Lignins. Tetrahedron Lett 36, 169–170, https://doi.org/10.1016/0040-4039(94)02203-N (1995).
Argyropoulos, D. S. et al. Abundance and reactivity of dibenzodioxocins in softwood lignin. J Agr Food Chem 50, 658–666, https://doi.org/10.1021/jf010909g (2002).
Zhang, L. M., Gellerstedt, G., Ralph, J. & Lu, F. C. NMR studies on the occurrence of spirodienone structures in lignins. J Wood Chem Technol 26, 65–79, https://doi.org/10.1080/02773810600580271 (2006).
Lagerquist, L. et al. Structural characterization of birch lignin isolated from a pressurized hot water extraction and mild alkali pulped biorefinery process. Industrial Crops and Products 111, 306–316, https://doi.org/10.1016/j.indcrop.2017.10.040 (2018).
Capanema, E. A., Balakshin, M. Y. & Kadla, J. F. A comprehensive approach for quantitative lignin characterization by NMR spectroscopy. J Agric Food Chem 52, 1850–1860, https://doi.org/10.1021/jf035282b (2004).
Shrivastava, A. In Introduction to Plastics Engineering (ed Anshuman Shrivastava) 17–48 (William Andrew Publishing, 2018).
Loh, X. J., Kai, D. & Li, Z. Functional Materials from Lignin. Vol. Volume 3 (WORLD SCIENTIFIC (EUROPE) 2017).
Heap, B. R. Permutations by Interchanges. The Computer Journal 6, 293–298, https://doi.org/10.1093/comjnl/6.3.293 (1963).
Hirayama, H. et al. Variation of the contents of biphenyl structures in lignins among wood species. 73, 569–578, https://doi.org/10.1515/hf-2018-0196 (2019).
Crestini, C., Melone, F., Sette, M. & Saladino, R. Milled wood lignin: a linear oligomer. Biomacromolecules 12, 3928–3935, https://doi.org/10.1021/bm200948r (2011).
Berinde, Z. & Berinde, M. On a matrix representation of molecular structures. Carpathian Journal of Mathematics 20, 205–209 (2004).
Weininger, D. Smiles, a Chemical Language and Information-System .1. Introduction to Methodology and Encoding Rules. J Chem Inf Comp Sci 28, 31–36, https://doi.org/10.1021/ci00057a005 (1988).
Willighagen, E. L. et al. The Chemistry Development Kit (CDK) v2.0: atom typing, depiction, molecular formulas, and substructure searching. J Cheminformatics 9, ARTN 3310.1186/s13321-017-0220-4 (2017).
te Velde, G. et al. Chemistry with ADF. Journal of Computational Chemistry 22, 931–967, https://doi.org/10.1002/jcc.1056 (2001).
Hanwell, M. D. et al. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminformatics 4, 17, https://doi.org/10.1186/1758-2946-4-17 (2012).
Pezoa, F., Reutter, J. L., Suarez, F., Ugarte, M. & Vrgoč, D. In Proceedings of the 25th International Conference on World Wide Web 263–273 (International World Wide Web Conferences Steering Committee, Montréal, Québec, Canada, 2016).
Probst, D. & Reymond, J.-L. A probabilistic molecular fingerprint for big data settings. J Cheminformatics 10, 66, https://doi.org/10.1186/s13321-018-0321-8 (2018).
Jmol: an open-source Java viewer for chemical structures in 3D, http://www.jmol.org/ (2022).
Zakzeski, J., Bruijnincx, P. C., Jongerius, A. L. & Weckhuysen, B. M. The catalytic valorization of lignin for the production of renewable chemicals. Chem Rev 110, 3552–3599, https://doi.org/10.1021/cr900354u (2010).
Gani, T. Z. H. et al. Computational Evidence for Kinetically Controlled Radical Coupling during Lignification. Acs Sustain Chem Eng 7, 13270–13277, https://doi.org/10.1021/acssuschemeng.9b02506 (2019).
Sander, T., Freyss, J., von Korff, M. & Rufener, C. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55, 460–473, https://doi.org/10.1021/ci500588j (2015).
Chakravarti, S. K. Distributed Representation of Chemical Fragments. Acs Omega 3, 2825–2836, https://doi.org/10.1021/acsomega.7b02045 (2018).
van der Maaten, L. Accelerating t-SNE using Tree-Based Algorithms. J Mach Learn Res 15, 3221–3245 (2014).
Alvarez-Vasco, C. et al. Unique low-molecular-weight lignin with high purity extracted from wood by deep eutectic solvents (DES): a source of lignin for valorization. Green Chem 18, 5133–5141, https://doi.org/10.1039/c6gc01007e (2016).
Donaldson, L. A. Lignification and lignin topochemistry - an ultrastructural view. Phytochemistry 57, 859–873, https://doi.org/10.1016/s0031-9422(01)00049-8 (2001).
Calvo-Flores, F. G., Dobado, J., Isac-García, J. & Martin-Martinez, F. Lignin and Lignans as Renewable Raw Materials: Biosynthesis of Lignin. (2015).
Rinaldi, R. et al. Paving the Way for Lignin Valorisation: Recent Advances in Bioengineering, Biorefining and Catalysis. Angew Chem Int Edit 55, 8164–8215, https://doi.org/10.1002/anie.201510351 (2016).
More, A., Elder, T. & Jiang, Z. H. A review of lignin hydrogen peroxide oxidation chemistry with emphasis on aromatic aldehydes and acids. Holzforschung 75, 806–823, https://doi.org/10.1515/hf-2020-0165 (2021).
Acknowledgements
The research described in this paper was partially supported by Pacific Northwest National Laboratory which is operated by Battelle Memorial Institute for the Department of Energy under contract DEAC05-76RLO1830. The authors are grateful for the support from the National Science Foundation [grant number 1454575] USDA NIFA [Award Number 2018-67009-27902] for funding part of this work. SCDE acknowledges the support received from the Pacific Northwest National Laboratory - Washington State University Distinguished Graduate Research Program and the U.S. Department of Energy BioEnergy Technologies Office under DEAC0576RL0-1830. We would like to thank Dr. Dylan Cronin and Dr. Kuan-Ting Lin from Washington State University for insightful discussions on the concept and Dr. Thomas Elder from USDA-Forest Service for his comments and suggestions to this paper.
Author information
Authors and Affiliations
Contributions
Eswaran S.C.D.: Formal analysis, Investigation, Model development and validation, Writing & Editing, Subramaniam S: Formal analysis, Editing & Review, Sanyal U: Investigation, validation and Review, Rallo R: Validation, Review & Editing, Proof reading, Supervision, Zhang, X: Conceptualize, Validation, Review & Editing, Supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41597_2022_1709_MOESM1_ESM.pdf
[Supplementary Material] Molecular structural dataset of lignin macromolecule elucidating experimental structural compositions
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Eswaran, S.c.d., Subramaniam, S., Sanyal, U. et al. Molecular structural dataset of lignin macromolecule elucidating experimental structural compositions. Sci Data 9, 647 (2022). https://doi.org/10.1038/s41597-022-01709-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-022-01709-4
This article is cited by
-
Effective Removal of Aqueous Lead (II) Ions Using Partially Esterified Lignin as a New Adsorbent
Iranian Journal of Science (2023)