Abstract
The Gutenberg Gait Database comprises data of 350 healthy individuals recorded in our laboratory over the past seven years. The database contains ground reaction force (GRF) and center of pressure (COP) data of two consecutive steps measured - by two force plates embedded in the ground - during level overground walking at self-selected walking speed. The database includes participants of varying ages, from 11 to 64 years. For each participant, up to eight gait analysis sessions were recorded, with each session comprising at least eight gait trials. The database provides unprocessed (raw) and processed (ready-to-use) data, including three-dimensional GRF and two-dimensional COP signals during the stance phase. These data records offer new possibilities for future studies on human gait, e.g., the application as a reference set for the analysis of pathological gait patterns, or for automatic classification using machine learning. In the future, the database will be expanded continuously to obtain an even larger and well-balanced database with respect to age, sex, and other gait-specific factors.
Measurement(s) | ground reaction force • centre of pressure • walking behavior • gait measurement • Normal Gait |
Technology Type(s) | force plate • Sensor Device |
Factor Type(s) | age • sex • walking speed |
Sample Characteristic - Organism | Homo sapiens |
Sample Characteristic - Environment | laboratory environment |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.15112488
Similar content being viewed by others
Background & Summary
The ability to walk is crucial for human mobility and is closely related to quality of life independent of age and sex1,2,3,4. The fear of losing the ability to walk is often considered as the most important concern of people after an accident or diagnosis, such as stroke5 or Parkinson’s disease6,7, and emphasizes the importance of walking for self-determined everyday life. In the healthcare sector, great efforts are made to prevent, diagnose, and rehabilitate limitations or even loss of independence due to gait impairments1,3,8. Three-dimensional instrumented gait analysis (3DGA) using video- or infrared-based motion capture systems and force plates is frequently used to objectively and quantitatively describe human locomotion. Consequently, 3DGA supports clinicians, therapists, and researchers in the standardized assessment of gait deviations and the detection of changes caused by orthopedic or physiotherapeutic interventions9,10. An evaluation using instrumented gait analysis is frequently accompanied by a large amount of data8,11,12, which are difficult to comprehend due to their multi-dimensional and multi-correlated nature13,14,15. The interpretation of such data can be a challenge even for experienced clinicians. Therefore, different approaches have been developed in recent years to facilitate the generation of meaningful clinical conclusions from 3DGA data and to support decision-making of clinical experts. Such approaches are based on, e.g., gait indexes15, multivariate statistical analysis13, and machine learning (ML)8,11,12,16. The latter are able to take into account and combine several time-continuous gait variables at once. These approaches can also support more experienced clinicians, whose evaluations are often based on subjective experiences with specific patient groups, by providing an objective perspective on the data.
In recent years, several ML-based approaches have been published that can assist clinicians in identifying individual gait characteristics17,18 and classifying specific gait patterns into clinically relevant categories16,19, e.g., stroke20, Parkinson’s disease21, cerebral palsy22, or specific functional gait disorders23. Although previous ML-based approaches provided promising results with respect to classification accuracy, these models have so far often been trained and evaluated on relatively small and well-controlled datasets as well as applied to simple classification tasks (e.g., healthy controls vs. Parkinson’s disease). The question of whether it is possible to train ML models that meet clinical requirements in terms of robustness, transparency, and generalizability has rarely been investigated. This has so far hindered broader clinical application and acceptance of ML models. The availability of sufficient and high-quality data is an important prerequisite for the training of reliable ML models. However, the availability of 3DGA data is often a limitation in practice. Among other authors24,25, we also made 3DGA data available to the public26,27,28,29,30 in previous studies17,31,32,33,34. However, different data processing procedures and data structures were used in these studies, making collaborative use of the data difficult. In recent years, a rather small number of annotated large-scale datasets have been made publicly accessible35. Publicly available datasets can be used to train more robust models. In practice, even with a large-scale dataset, such as GaitRec35, data from individuals without gait pathology (healthy controls) represent a bottleneck. One reason for the scarcity of such data is that most gait analysis laboratories are located at clinics and usually record and examine only patients with pathological gait patterns.
In order to address this shortcoming, we provide - with the Gutenberg Gait Database - the gait data from healthy controls collected in our laboratory over the past seven years. The data is provided in a uniform format to allow for a continuously growing and publicly accessible database. The overall goal is to bridge existing gaps in publicly available gait datasets. Thereby, we aim at creating a basis for reliable ML models that can be used as decision-support system in clinical practice and research. Based on this goal, we prepared the processed data in such a way that it can be merged and used in conjunction with the GaitRec dataset35. In addition, the size and quality of the database allow it to serve as an extension of the study population in gait-related research areas, e.g., shoe and insole research36, security systems based on biometric recognition37, gait-based fatigue38 and emotion39 detection in psychological and sport-related contexts. In this setting our database can be used in various ways, e.g., as reference data or as source for automatic outlier detection. From a more epistemological point of view, the continuously growing database will also allow increasing flexibility in dealing with much more diverse questions related to human gait. Questions concerning population-motivated research40, problems of specific groups41, or the complexity of individual case-oriented time series31,32 will be put on a broader data-based foundation over time.
The Gutenberg Gait Database provides exclusively force plate data, namely ground reaction force (GRF) and center of pressure (COP) signals. The current best practice in clinical gait analysis describes a patient’s gait using a combination of force plate data with kinematic and electromyographic data. However, kinematic and electromyographic data are prone to several difficulties, such as inconsistencies due to differences in anthropometric characteristics of participants, experience of investigators, measurement protocols, and laboratory settings42,43,44. This makes it more difficult to create a homogeneous, large-scale, and high-quality dataset compared to using less interference-prone data, such as GRF signals45,46. Therefore, the use of force plate data offers advantages for the development of ML models for gait analysis, although the provided information appears to be reduced in comparison to kinematic data. However, previous studies23,47 investigating ML methods for automated classification of gait impairments based on force plate data showed promising results suggesting their suitability for clinical applications.
Methods
Datasets
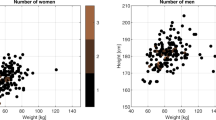
The Gutenberg Gait Database combines datasets from five already published studies on human gait17,31,32,33,34 and data from five unpublished studies. A total sample of 350 participants (142 female, 205 male, and 3 unknown) aged between 11 and 64 years is included. Prior to the recording, all participants reported that they did not have any gait pathology and were not suffering from any injuries or diseases that affected gait. Table 1 summarizes demographic details for each individual dataset and the total database. Figure 1 shows the overall and sex-specific distributions of age, body mass, body height, and walking speed for the database.
Frequency distribution of age, body mass, body height, and walking speed for all (upper panel), female (middle panel), and male (lower panel) participants. The distributions are based on the values of the initial session of each participant. For the waking speed, the mean values of the gait trials of the initial session are shown.
All studies (published and unpublished) were carried out according to the Declaration of Helsinki at the Johannes Gutenberg-University in Mainz (Germany). All participants were informed about the experimental protocol and provided their written informed consent to participate in the study. The approval from the ethical committee of the medical association Rhineland-Palatinate in Mainz (Germany) was received.
Data recording & Experimental protocol
Bi-lateral analog force plate signals were recorded by asking participants to walk at their preferred (self-selected) walking speed on a level and approximately 10 m long walkway. Two force plate configurations were used: (i) an inline configuration using two centrally embedded force plates (Kistler, Type 9287CA, Switzerland) and (ii) a staggered configuration using two force plates (Kistler, Type 9286AA, Switzerland) integrated in a wooden walkway.
For both force plate configurations, the analog force plate signals were amplified (Kistler, Type 5233 A, Switzerland) and converted to digital signals using a sampling frequency of 1,000 Hz. A data acquisition system (Kistler, Type 5695, Switzerland) with a 16-bit analog-digital converter (Measurement Computing Corporation, Type USB-2533, USA) was used with a signal input range of ±10 V. Depending on the underlying experimental protocol, the walking speed was either estimated using (i) two light barriers with two photoelectric sensors (Imhof Timing, Germany) at a sampling frequency of 1,000 Hz or (ii) the three-dimensional pelvis marker trajectories captured by nine infrared cameras (Qualisys AB, Type Oqus 310, Sweden) at a sampling frequency of 250 Hz.
Participants were asked to perform gait trials to familiarize with the experimental setup and to determine an individual starting position for the gait analysis session. The number of familiarization trials differed between the experimental protocols. The exact number is specified for each study in Table 2. This procedure has already been shown to minimize the impact of targeting the force plates on the observed gait variables48,49. In addition, the participants were instructed to look at a symbol (neutral smiley) on the opposing wall of the laboratory to direct their attention away from the force plates and ensure a natural walk with an upright body position.
During one gait analysis session, participants walked until a predefined number of valid gait trials were available. These gait trials were defined as valid by the assessor if the participant walked “naturally” (e.g., with respect to force plate targeting) and both force plates were hit cleanly. The predefined number of gait trials per session varied between the experimental protocols and ranged from 8 to 20 gait trials. The exact number for each experimental protocol is specified in Table 2. Depending on the experimental study design, one to eight gait analysis sessions were recorded per participant.
Data processing
The three-dimensional GRFs (vertical, anterior-posterior, and medio-lateral) and the two-dimensional COPs (anterior-posterior and medio-lateral) were calculated based on the analog force plate signals. The database provides unprocessed (raw) and processed (ready-to-use) GRF and COP signals during the stance phase. The data processing procedure was coordinated with Horsak et al.35 so that the processing of the data in the Gutenberg Gait Database is identical to the GaitRec dataset. Thereby, we were able to prevent the obstacles that often exist in practice when using different datasets jointly. The main benefit for the community is the combined use of both data sources. We have, thus, eliminated a major disadvantage of the GaitRec dataset, namely ensuring that the number of healthy control participants is no longer a bottleneck.
For both settings, i.e., unprocessed and processed data, following pre-processing steps were performed. The offset of each analog force plate signal was corrected using the mean value of the first ten frames. The analog force plate signals were down-sampled to 250 Hz. The orientation of the medio-lateral and anterior-posterior GRF and COP signals were unified. Thus, medial and anterior forces were transformed to positive and lateral and posterior to negative values.
For the unprocessed (raw) data, we determined the signals in the following way. The stance phase was determined using a vertical GRF threshold of 25 N. The cropped GRF signals of the stance phase were used to calculate the COP signals.
For the processed (ready-to-use) data, we filtered the GRF signals using a second-order Butterworth bidirectional low-pass filter at a cut-off frequency of 20 Hz. The stance phase was determined based on the filtered GRF signals using a vertical GRF threshold of 25 N. For the processed COP signals, we filtered the unprocessed (raw) COP signals as well with a second-order Butterworth bidirectional low-pass filter at a cut-off frequency of 20 Hz. Furthermore, we cropped the filtered COP signals with a vertical GRF threshold of 80 N to avoid artifacts in COP calculation at small GRF signal values. In addition, the medio-lateral COP signals were mean-centered and anterior-posterior COP signals zero-centered. Each GRF and COP signal was time-normalized to 101 data points, corresponding to 100% stance phase. The GRF signals were normalized to the body weight, measured before each gait analysis session. The whole data processing was performed within the Matlab 2019a (The MathWorks, Inc., Natick, Massachusetts, USA) framework.
Data Records
All published data are fully anonymized and are available online from figshare50. As already pointed out, we decided to follow the data processing procedure and data structure as well as the naming of the files according to the GaitRec dataset35. The data records consist of twenty files containing the GRF data for each gait trial (see Table 3) and one file containing the measured walking speed for each gait trial. In addition, we provide one file containing metadata for each gait analysis session, including additional participants’ information, e.g., class label, sex, age, body mass. All files are available as comma-separated value files (.csv). The twenty GRF data files are organized according to the following naming convention: “GRF-type-processing-side.csv”. The type denotes, whether the file holds the vertical (“F_V”), anterior-posterior (“F_AP”), medio-lateral (“F_ML”) or the anterior-posterior or medio-lateral COP (“COP_AP”, “COP_ML”) time-series. Processing denotes, if the files hold the unprocessed (raw) data (“RAW”) or the processed (ready-to-use) data (“PRO”). The side denotes, if the data are from the “left” or “right” body side. The common prefix for all files is “GRF-”. An example filename is: “GRF_F_V_RAW_left.csv”.
Each of the “GRF-type-processing-side.csv” files is structured as a matrix with T rows × K columns (T = 8,819; K = 105 for “PRO” and K = 216 for “RAW”). Each row holds the data of one gait trial. The first column identifies each dataset (“DATASET_ID”), the second column each participant (“SUBJECT_ID”), the third column each gait analysis session (“SESSION_ID”), and the fourth column each single gait trial within a session (“TRIAL_ID”). The remaining columns contain the values of the GRF signals for each gait trial. Note that due to the non-normalized nature of the data and the resulting different time-series lengths in the “RAW” files, non-available numbers have been replaced by “NaN” to maintain a constant matrix-dimension.
The file holding the measured walking speed for each gait trial is named “GRF_walking_speed.csv”. The file is structured as a matrix with T rows × L columns (T = 8,819; L = 5). Each row holds the data of one gait trial. The first column identifies each dataset (“DATASET_ID”), the second column each participant (“SUBJECT_ID”), the third column each gait analysis session (“SESSION_ID”), and the fourth column each single gait trial within a session (“TRIAL_ID”). The fifth column contains the measured walking speed for each gait trial (“WALKING_SPEED”). The walking speed was not measured in datasets 6, 8, and 10. Non-available numbers have been replaced by “NaN” to maintain a constant matrix-dimension.
The metadata file, which contains additional participant and session-related information is named “GRF_metadata.csv” (see Table 4). The file is structured as a matrix with S rows × M columns (S = 661; M = 21). Here, the first three columns hold the DATASET_ID, SUBJECT_ID, and SESSION_ID, the other columns hold information such as sex, body mass, and age (see Table 4 for more details). Non-available numbers have been replaced by “NaN” to maintain a constant matrix-dimension.
Technical Validation
The force plates and the measurement equipment were calibrated by the manufacturer (Kistler, Switzerland) and regularly checked and serviced during laboratory practice. No specific procedure (e.g., such as the CalTester method) was used.
In addition, on each day when measurements were conducted, the proper functioning of the force plates and measuring equipment was ensured by the following procedure: (i) A 30 s recording without load on the force plates was taken and ensured that the signal noise was below ±1 N. (ii) The assessor performed a weight measurement to verify the proper amplification of the analog channels. (iii) The assessor walked along the 10 m analysis walkway with one foot contact on each force plate and verified that the GRF signals showed the characteristic curves.
For an impression of data integrity, the processed data for each dataset is shown in Fig. 2 (GRF) and Fig. 3 (COP).
Visualization of vertical (left panel), anterior-posterior (central panel), and medio-lateral (right panel) force components of the body weight (BW)-normalized GRF measurements per dataset. Mean and standard deviation signals (calculated per dataset) are highlighted as solid and dashed colored lines.
Visualization of zero-centered anterior-posterior (left panel) and mean-centered medio-lateral (right panel) components of the COP measurements per dataset. Mean and standard deviation signals (calculated per dataset) are highlighted as solid and dashed colored lines. We carefully inspected the gait trials where the signals differed considerably and made sure that these differences were not the result of measurement or calculation errors. Using the kinematic data, we were able to verify that the deviating signals were from gait trials of forefoot or midfoot walking participants.
Usage Notes
The data are stored in *.csv files and can be easily imported into any software framework for further data analysis. We provide two scripts that allow a straightforward data import for Matlab (The MathWorks, Inc., Natick, Massachusetts, United States, 2019a) and Python (Python Software Foundation, 3.7). Additionally, two scripts (for Matlab and Python) are available for merging the GaitRec dataset35 and the Gutenberg Gait Database. For the GaitRec dataset the DATASET_ID is set to 0. Since the metadata files and the data files have the same structure, a simple consolidation can be achieved. The GaitRec dataset has a bottleneck in terms of healthy control participants. Merging the two datasets can compensate for this limitation and allow the data to be much more useful for future research. Merging the two data sources would increase the number of healthy controls from 211 to 561, which approximately corresponds to the cardinality of the gait disorder classes: hip (N = 450), knee (N = 625), ankle (N = 627), calcaneus (N = 382).
Code availability
A custom script for tracing and replicating the used processing of the force plate data in Matlab (The MathWorks, Inc., Natick, Massachusetts, United States, 2019a) and custom scripts for importing and merging (with the GaitRec dataset) the data in Matlab (The MathWorks, Inc., Natick, Massachusetts, United States, 2019a) and Python (Python Software Foundation, 3.7) are publicly available at figshare50.
References
Verghese, J. et al. Epidemiology of gait disorders in community-residing older adults. J. Am. Geriat. Soc. 54, 255–261, https://doi.org/10.1111/j.1532-5415.2005.00580.x (2006).
Fagerström, C. & Borglin, G. Mobility, functional ability and health-related quality of life among people of 60 years or older. Aging Clin. Exp. Res. 22, 387–394, https://doi.org/10.1007/BF03324941 (2010).
Mahlknecht, P. et al. Prevalence and burden of gait disorders in elderly men and women aged 60-97 years: A population-based study. PloS one 8, e69627, https://doi.org/10.1371/journal.pone.0069627 (2013).
Forte, R., Boreham, C. A. G., de Vito, G. & Pesce, C. Health and quality of life perception in older adults: The joint role of cognitive efficiency and functional mobility. Int. J. Environ. Res. Public Health 12, 11328–11344, https://doi.org/10.3390/ijerph120911328 (2015).
Schmid, A. et al. Improvements in speed-based gait classifications are meaningful. Stroke 38, 2096–2100, https://doi.org/10.1161/STROKEAHA.106.475921 (2007).
Ellis, T. et al. Which measures of physical function and motor impairment best predict quality of life in Parkinson’s disease? Parkinsonism Relat. Disord. 17, 93–697, https://doi.org/10.1016/j.parkreldis.2011.07.004 (2011).
Soh, S. E., Morris, M. E. & McGinley, J. L. Determinants of health-related quality of life in Parkinson’s disease: A systematic review. Parkinsonism Relat. Disord. 17, 1–9, https://doi.org/10.1016/j.parkreldis.2010.08.012 (2015).
Prakash, C., Kumar, R. & Mittal, N. Recent developments in human gait research: parameters, approaches, applications, machine learning techniques, datasets and challenges. Artif. Intell. Rev. 49, 1–40, https://doi.org/10.1007/s10462-016-9514-6 (2018).
Baker, R. J. Measuring Walking: A Handbook Of Clinical Gait Analysis (Mac Keith Press, London, 2013).
Wren, T. A. L., Tucker, C. A., Rethlefsen, S. A., Gorton, G. E. & Õunpuu, S. Clinical efficacy of instrumented gait analysis: Systematic review 2020 update. Gait Posture 80, 274–279, https://doi.org/10.1016/j.gaitpost.2020.05.031 (2020).
Phinyomark, A., Petri, G., Ibáñez-Marcelo, E., Osis, S. T. & Ferber, R. Analysis of big data in gait biomechanics: Current trends and future directions. J. Med. Biol. Eng. 38, 244–260, https://doi.org/10.1007/s40846-017-0297-2 (2018).
Halilaj, E. et al. Machine learning in human movement biomechanics: Best practices, common pitfalls, and new opportunities. J. Biomech. 81, 1–11, https://doi.org/10.1016/j.jbiomech.2018.09.009 (2018).
Chau, T. A review of analytical techniques for gait data. Part 1: Fuzzy, statistical and fractal methods. Gait Posture 13, 49–66, https://doi.org/10.1016/S0966-6362(00)00094-1 (2001).
Wolf, S. et al. Automated feature assessment in instrumented gait analysis. Gait Posture 23, 331–338, https://doi.org/10.1016/j.gaitpost.2005.04.004 (2006).
Cimolin, V. & Galli, M. Summary measures for clinical gait analysis: A literature review. Gait Posture 39, 1005–1010, https://doi.org/10.1016/j.gaitpost.2014.02.001 (2014).
Schöllhorn, W. I. Applications of artificial neural nets in clinical biomechanics. Clin. Biomech. 19, 876–898, https://doi.org/10.1016/j.clinbiomech.2004.04.005 (2004).
Horst, F., Lapuschkin, S., Samek, W., Müller, K.-R. & Schöllhorn, W. I. Explaining the unique nature of individual gait patterns with deep learning. Sci. Rep. 9, 2391, https://doi.org/10.1038/s41598-019-38748-8 (2019).
Aeles, J., Horst, F., Lapuschkin, S., Lacourpaille, L. & Hug, F. Revealing the unique features of each individual’s muscle activation signatures. J. R. Soc. Interface 18, 20200770, https://doi.org/10.1098/rsif.2020.0770 (2021).
Figueiredo, J., Santos, C. P. & Moreno, J. C. Automatic recognition of gait patterns in human motor disorders using machine learning: A review. Med. Eng. Phys. 53, 1–12, https://doi.org/10.1016/j.medengphy.2017.12.006 (2018).
Lau, H.-y, Tong, K.-y & Zhu, H. Support vector machine for classification of walking conditions of persons after stroke with dropped foot. Hum. Mov. Sci. 28, 504–514, https://doi.org/10.1016/j.humov.2008.12.003 (2009).
Wahid, F., Begg, R. K., Hass, C. J., Halgamuge, S. & Ackland, D. C. Classification of parkinson’s disease gait using spatial-temporal gait features. IEEE J. Biomed. Health Inform. 19, 1794–1802, https://doi.org/10.1109/JBHI.2015.2450232 (2015).
Van Gestel, L. et al. Probabilistic gait classification in children with cerebral palsy: A bayesian approach. Res. Dev. Disabil. 32, 2542–2552, https://doi.org/10.1016/j.ridd.2011.07.004 (2011).
Slijepcevic, D. et al. Automatic classification of functional gait disorders. IEEE J. Biomed. Health Inform. 22, 1653–1661, https://doi.org/10.1109/JBHI.2017.2785682 (2018).
Fukuchi, C. A., Fukuchi, R. K. & Duarte, M. A public dataset of overground and treadmill walking kinematics and kinetics in healthy individuals. PeerJ 6, e4640, https://doi.org/10.7717/peerj.4640 (2018).
Schreiber, C. & Moissenet, F. A multimodal dataset of human gait at different walking speeds established on injury-free adult participants. Sci. Data 6, 111, https://doi.org/10.1038/s41597-019-0124-4 (2019).
Horst, F. et al. A public dataset of overground walking kinetics and lower-body kinematics in healthy adult individuals on different days. Mendeley Data https://doi.org/10.17632/8kyv4jm759.1 (2019).
Horst, F., Eekhoff, A., Newell, K. M. & Schöllhorn, W. I. A public dataset of overground walking kinetics and lower-body kinematics in healthy adult individuals on different sessions within one day. Mendeley Data https://doi.org/10.17632/b48n46bfry.1 (2019).
Horst, F., Mildner, M. & Schöllhorn, W. I. A public dataset of overground walking kinetics in healthy individuals. Mendeley Data https://doi.org/10.17632/yrpbd8fhc4.1 (2018).
Horst, F., Lapuschkin, S., Samek, W., Müller, K.-R. & Schöllhorn, W. I. A public dataset of overground walking kinetics and full-body kinematics in healthy individuals. Mendeley Data https://doi.org/10.17632/svx74xcrjr.3 (2019).
Burdack, J. et al. A public dataset of overground walking kinetics in healthy adult individuals on different sessions within one day. Mendeley Data https://doi.org/10.17632/y55wfcsrhz.2 (2020).
Horst, F. et al. Daily changes of individual gait patterns identified by means of support vector machines. Gait Posture 49, 309–314, https://doi.org/10.1016/j.gaitpost.2016.07.073 (2016).
Horst, F., Mildner, M. & Schöllhorn, W. I. One-year persistence of individual gait patterns identified in a follow-up study - A call for individualised diagnose and therapy. Gait Posture 58, 476–480, https://doi.org/10.1016/j.gaitpost.2017.09.003 (2017).
Horst, F., Eekhoff, A., Newell, K. M. & Schöllhorn, W. I. Intra-individual gait patterns across different time-scales as revealed by means of a supervised learning model using kernel-based discriminant regression. PloS one 12, e0179738, https://doi.org/10.1371/journal.pone.0179738 (2017).
Burdack, J. et al. Systematic Comparison of the Influence of Different Data Preprocessing Methods on the Performance of Gait Classifications Using Machine Learning. Front. Bioeng. Biotechnol. 8, 260, https://doi.org/10.3389/fbioe.2020.00260 (2020).
Horsak, B. et al. GAITREC: A large-scale ground reaction force dataset of healthy and impaired gait. Sci. Data 7, 143, https://doi.org/10.1038/s41597-020-0481-z (2020).
Nigg, B. M. Biomechanics Of Sport Shoes (University of Calgary, Calgary, 2010).
Mason, J.E. Traoré, I. & Woungang, I. Machine Learning Techniques For Gait Biometric Recognition. (Springer International Publishing, Basel, 2016).
Janssen, D. et al. Diagnosing Fatigue in Gait Patterns by Support Vector Machines and Self-organizing Maps. Hum. Mov. Sci. 5, 966–975, https://doi.org/10.1016/j.humov.2010.08.010 (2011).
Janssen, D. et al. Recognition of Emotions in Gait Patterns by Means of Artificial Neural Nets. J. Nonverbal Behav. 32, 79–92, https://doi.org/10.1007/s10919-007-0045-3 (2008).
Vuillermin, C. et al. Severe crouch gait in spastic diplegia can be prevented: a population-based study. J. Bone Joint Surg. Br. 93, 1670–1675, https://doi.org/10.1302/0301-620X.93B12.27332 (2011).
Simonsen, E. B. & Alkjær, T. The Variability Problem of Normal Human Walking. Med. Eng. Phys. 34, 219–224, https://doi.org/10.1016/j.medengphy.2011.07.013 (2012).
Schwartz, M. H., Trost, J. P. & Wervey, R. A. Measurement and management of errors in quantitative gait data. Gait Posture 20, 196–203, https://doi.org/10.1016/j.gaitpost.2003.09.011 (2004).
Gorton, G. E., Hebert, D. A. & Gannotti, M. E. Assessment of the kinematic variability among 12 motion analysis laboratories. Gait Posture 29, 398–402, https://doi.org/10.1016/j.gaitpost.2008.10.060 (2009).
McGinley, J. L., Baker, R. J., Wolfe, R. & Morris, M. E. The reliability of three-dimensional kinematic gait measurements: A systematic review. Gait Posture 29, 360–369, https://doi.org/10.1016/j.gaitpost.2008.09.003 (2009).
Kadaba, M. P. et al. Repeatability of kinematic, kinetic, and electromyographic data in normal adult gait. J. Orthop. Res. 7, 849–860, https://doi.org/10.1002/jor.1100070611 (1989).
Benedetti, M. G., Merlo, A. & Leardini, A. Inter-laboratory consistency of gait analysis measurements. Gait Posture 38, 934–939, https://doi.org/10.1016/j.gaitpost.2013.04.022 (2013).
Alaqtash, M., et al Automatic classification of pathological gait patterns using ground reaction forces and machine learning algorithms. In 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS), 453–457, https://doi.org/10.1109/IEMBS.2011.6090063 (IEEE, 2011).
Wearing, S. C., Urry, S. R. & Smeathers, J. E. The effect of visual targeting on ground reaction force and temporospatial parameters of gait. Clin. Biomech. 15, 583–591, https://doi.org/10.1016/s0268-0033(00)00025-5 (2000).
Sanderson, D. J., Franks, I. M. & Elliott, D. The effects of targeting on the ground reaction forces during level walking. Hum. Mov. Sci. 12, 327–337, https://doi.org/10.1016/0167-9457(93)90022-H (1993).
Horst, F., Slijepcevic, D., Simak, M. & Schöllhorn, W. I. Gutenberg Gait Database: A ground reaction force database of level overground walking in healthy individuals. figshare https://doi.org/10.6084/m9.figshare.c.5311538 (2021).
Acknowledgements
This work was supported by the internal research grant “inneruniversitäre Forschungsförderung” from the Johannes Gutenberg-University Mainz as well as the Lower Austrian Research and Education Company (NFB) and the Provincial Government of Lower Austria (IntelliGait3D – #FTI17-014). The authors thank David Corell, Sabrina Daffner, Alexander Eekhoff, Ibrahim Hassan, Patrick Hegen, Eva Klein, Franziska Kramer, Kathrin Kronemayer-Wurm, Markus Mildner, Christin Rupprecht, Bastian Schäfer, and Nathalie Scherdel for their encouragement and support during data collection.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
F.H. and W.I.S. raised funding for this work. F.H. and M.S. prepared the datasets. F.H., D.S. and M.S. processed the data. D.S. created the data files and implemented the import scripts. F.H. and D.S. wrote the manuscript. D.S. designed the figures. F.H., D.S., M.S. and W.I.S. reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Horst, F., Slijepcevic, D., Simak, M. et al. Gutenberg Gait Database, a ground reaction force database of level overground walking in healthy individuals. Sci Data 8, 232 (2021). https://doi.org/10.1038/s41597-021-01014-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-01014-6
This article is cited by
-
Real-world measurements of ground reaction forces of normal gait of young adults wearing various footwear
Scientific Data (2023)
-
A Multi-Modal Gait Database of Natural Everyday-Walk in an Urban Environment
Scientific Data (2022)