Abstract
Tree provenance trials consist of a variety of seed sources (or provenances) planted at several test sites across the range of a species. The resulting plantations are typically measured periodically to investigate provenance performance in relation to abiotic conditions, particularly climate. These trials are expensive and time consuming to establish, but are an important resource for seed transfer systems, which aim to match planting sites with well-adapted (climatically suitable) seed sources. Provenance trial measurements may be underutilized because the data are scattered across publications, conference proceedings, and university theses. Here we document an effort to collect available provenance trial measurements and associated climate data for seven eastern North American tree species (Pinus strobus, Pinus banksiana, Picea glauca, Picea mariana, Quercus rubra, Larix laricina, Betula alleghaniensis). The resulting datasets included a total of 773 provenances and 62 test sites, with 65 historical climate variables appended to each location. We hope this data will support forest managers in making seed transfer decisions, particularly in an era of rapid climate change.
Measurement(s) | height • survival • phenology |
Technology Type(s) | digital curation |
Sample Characteristic - Organism | Pinus strobus • Pinus banksiana • Picea glauca • Picea mariana • Quercus rubra • Larix laricina • Betula alleghaniensis |
Sample Characteristic - Environment | woodland area |
Sample Characteristic - Location | Canada • United States of America |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.13136060
Similar content being viewed by others
Background & Summary
Natural populations of tree species often vary in traits such as cold hardiness, growth, and bud phenology across their geographic range, due to local adaptation1. Much research has been done to quantify the variation in these traits across a species’ range for the purpose of elucidating population-level preferences in environmental conditions. Generally, tree provenance studies involve collecting representative seed of natural populations from a variety of locations (also referred to as provenances) across the range of a species and subsequently planting them at one or more test sites with statistically sound experimental designs. The resulting plantations are monitored through time by measuring performance-related variables such as tree height and survival. Better adapted populations are expected to grow faster and exhibit higher survival rates. Although a common assumption is that the local provenance is best, this is not always the case. For example, Lu et al. showed that white spruce provenances from southern Ontario, Canada, performed better (in terms of height and survival) than local provenances at several northwestern Ontario test sites1. There are several potential explanations for the success of non-local provenances, including recent climate change and the migration history of the species1.
An important application of provenance data is in the development of seed transfer guidelines2. In the past, transfer guidelines have typically aimed to limit seed movements in order to ensure the use of local seed sources at reforestation sites. However, with an evolving climate, directional seed movements (usually poleward or upslope) may be needed to maintain a reasonable match between historical climate at the seed source and future climate at the planting site. Furthermore, optimizing future growth gains obtained by planting non-local provenances must be balanced with possible near-term mortality due to climate/weather-related factors such as winter injury. Since provenance trials gather information on provenance performance across a range of climatic conditions, they are well-suited to provide valuable insights into optimal seed transfer movements under climate change. Unfortunately, tree provenance trials are expensive to establish, maintain, and measure. Further, the long lifespan of trees means that it can take decades to obtain information about mature trees from provenance trials. For these reasons, consolidating existing provenance data into a publicly available repository is an important undertaking.
No current resource provides compiled data from eastern North American tree provenance trials in a single product with associated climate data. Instead, these data are scattered across a variety of sources, including journal articles, conference proceedings, university theses, and government reports. Here we present a data product that consolidates provenance data and associated climate estimates for seven major eastern North American tree species. This effort provides timely data to researchers and forest managers given the recent increased focus on seed transfer under climate change. Prospects for rapid climate change over the coming decades3 and management strategies such as assisted migration increase the relevance of historical data4. Climate and elevation data are included along with the provenance measurements, in order to, for example, facilitate the calculation of safe climate transfer distances5 for each tree species in the database. This article details the data acquisition and data processing methods used to create the databases and describes the structure of the databases and related quality assurance methods. We hope that the databases and information provided in this article will facilitate the use of existing tree provenance data for ongoing seed transfer research and decision making.
Methods
Study design & database considerations
We identified seven major tree species that have significant economic and/or ecological value in our geographic area of interest (northeastern United States and eastern Canada): Picea glauca (white spruce), Picea mariana (black spruce), Pinus banksiana (jack pine), Larix laricina (tamarack), Pinus strobus (white pine), Quercus rubra (red oak), and Betula alleghaniensis (yellow birch)). White spruce, black spruce, and jack pine are important commercial conifer species harvested in large volumes across the northern (boreal) portion of our area of interest6. Red oak, yellow birch, and white pine are high value hardwood species7,8 that have more southern geographic ranges, but have potential for future northward seed transfer. We included data for seed sources from across North America where available, although most originated in the northeastern United States and eastern Canada.
We gathered information for three types of measurement (or response) variables, including tree height, survival, and phenology. Tree height is widely used to indicate provenance growth potential in seed transfer research, but tree survival and phenology data are also important to indicate that a potential seed source can survive and adapt to the conditions at a certain location. Phenology data is of particular interest due to changes in seasonality associated with climate change and the lack of information on how species and populations will respond to these changes9. However, this data is not widely reported in tree provenance trials because it is time sensitive and challenging to measure, with multiple visits required to accurately record the phenological stage of the buds10.
In order to identify potential data sources and create a functional database that included both provenance and climate data, we completed six major steps (Fig. 1).
Systemic review, metadata collection, & selection of sources
We conducted an extensive literature review of sources (publications, conference proceedings, government reports, and university theses) relating to tree provenance trials, for each of the seven tree species of interest. We used keywords such as “provenance trials”, “height growth”, “tree survival”, “common garden study”, and both the common and scientific names of the tree species when searching for sources using resources such as Google Scholar and Reforestation, Nurseries, & Genetic Resources (RNGR). We reviewed a total of 130 sources, recording metadata such as the number of test sites, the year an individual trial was established, the number of provenances included in each trial, provenance mean values of the variables measured in individual trials, tree age at measurement, etc. We also recorded how trees in each trial were measured for each response variable and whether the trial results were reported in-text. If the location information and trial data were reported and it was likely that all trees in the trials were measured, we digitized the information related to the study in a series of tables. Online-only Table 1 provides a summary of sources selected for inclusion in our data product.
Data processing
All the records in the database were converted to consistent units, such as decimal degrees for latitude and longitude, meters for tree height, and days since January 1 for phenology data. All measurements included in the database represent the mean value for the whole test site, including any replications. For some studies, we assumed this to be the case because no information was provided in the original study about methods used to sample the trees to take measurements. In some cases, there were multiple test sites at one location, representing different environments11. In this case, the test sites were entered separately, labeled as “Site 1”, “Site 2”, etc. We also included the test sites separately for different species or studies.
Climate data associated with provenances and test sites
Estimates for some 65 climate variables (see Online-only Table 2 for more details) were obtained by interrogating existing spatial climate models at each seed source and test site location12. Growing season-related variables were calculated using a Bessel interpolation in conjunction with monthly climate variables13. The climate estimates included averages for the 1961–1990 period, which provide a reasonable approximation of historical long-term climate values at seed source locations. In addition, climate estimates were generated at each test site for each year over the period spanning plantation establishment and measurement. Specifically, we used a Python program to calculate mean, standard deviation, minimum, and maximum climate values over this period.
Once we generated climate estimates at each test site and seed source location, we used a Python script to append the information (using a unique seed source identification code as the primary key) to the tables containing the provenance trial data. If elevation information had not been reported in the original document, we appended elevation estimates at seed source and test site locations using a digital elevation model (DEM)14.
Data Records
Data format & storage location
The databases are provided in a series of comma-delimited files (available in both.txt and.csv format). These tables are organized in a series of folders (see Fig. 2). The first database (henceforth the trial results database) contains information on seed sources from 22 provenance studies in eastern North America, including latitude, longitude, elevation, performance measurements (tree height, survival, and phenology data), and 65 long-term climate variables (i.e., averages for the 1961–1990 period). The second database (henceforth the test sites database) contains information on test sites from the same 22 studies, including latitude, longitude, elevation, and 65 climate variables summarized for the period spanning plantation initiation and measurement. All data were recorded in standardized format and units (decimal degrees, meters, °C, etc.). The databases are accessible in an online repository at https://doi.org/10.5063/K072NQ15.
Here, we have provided two versions of the trial results database. The first version of the database (trial_results_database) contains tables with consistent columns to facilitate data analysis in environments such as R. If a certain test site did not have a specific measurement, the column was included but contained no data values. This database contains mean height and survival for all trial measurement ages in single columns (i.e., “Mean Height (m)”, “Survival (%)”). To achieve this, we entered multiple records for each seed source. The second version of the database (trial_results_database_primary_key) contains tables with a single row for each seed source to facilitate data analysis with SQL in environments such as Microsoft Access or PostgreSQL. In this case, we included individual columns for each measurement taken at each test site (i.e., “Mean Height Age 10 (m)”). This format means that the seed source identification code can be used as the primary key and can be used to join tables from the same study.
Data structure & citation
The trial results database is organized by species and contains a table for each test site, identified by both the name of the test site location and the data source number. This database also contains information about the data sources, such as the accuracy of the latitude and longitude of the seed sources (whether it was reported in-text or estimated from the location name) and whether or not phenology data was available. In total, this database contains 75 tables. Table 1 reports an overview of the information contained in this database. Detailed information on the data contained in each table can be found in the database, in the table “Table Overview”.
The test sites database is organized by test site and includes tables that contain mean, standard deviation, minimum, and maximum climate values for the period between plantation establishment and measurement. If there were multiple measurement years, additional tables were provided for the period between trial establishment and the later measurement years. For example, if a test site was established in 1975 and measured in 1979 and 1980, we included two tables, one for 1975–1979 and one for 1979–1980. In order to avoid tables in the database having the same name, the tables are also named with the unique code of the test site, such as 1988–1990 (pjt281). The purpose of this is to avoid errors when querying the database. The test site database also contains information about the data sources, as well as the test sites (such as whether the elevation data comes from the data source or the DEM). Missing data values were denoted using −9999. In total, this database contains 103 tables. Figure 2 shows a schematic of the folder structure of both databases.
Both databases are organized such that the source of the data is noted alongside the test site name. The full citations of the data sources are found in both databases, in the table named “Guide to Data Sources”. For users of the data, the original source should be cited along with the database.
Applications
The databases presented here will support seed transfer analyses. The provenance trial data, in combination with the associated climate variables, can be used to calculate critical seed transfer distances – i.e., the distance that a seed source can be moved to improve long term productivity but also minimize the risk of near term mortality16. The information provided in the databases may contribute to efforts to develop more robust transfer functions (a function to model climate-performance relationships) by including more test sites and/or seed sources17. Furthermore, the datasets can also be used for applications such as mapping trends and finding correlations between climate variables and trial results.
Technical Validation
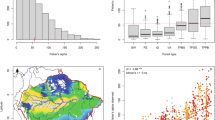
The first method we used for data quality assurance was to examine the data for unreasonable values. For example, tree height mean data for each test site was sorted in ascending and descending order to examine the data for any extreme values. Suspect values were then compared with the source data to check for data entry errors. For quality assurance of the climate data, we mapped the values for mean annual temperature for each provenance and test site in ArcMap (Fig. 3); similar efforts were undertaken for a selection of the other climate variables as well (Figs. 4, 5). Accuracy of the mean, standard deviation, minimum, and maximum calculations was ensured by comparing the results with hand calculations for a selection of data values to ensure that the Python program was working properly.
Code availability
We used various Python scripts to process the data for input into the databases. These scripts were used to calculate climate value summaries and convert phenology observations to a uniform reference date (January 1). The scripts are available at https://github.com/clara-risk/tree-provenance-trials.
References
Lu, P., Parker, W. C., Colombo, S. J. & Man, R. Restructuring tree provenance test data to conform to reciprocal transplant experiments for detecting local adaptation. J. Appl. Ecol. 53, 1088–1097 (2016).
Pedlar, J. H. et al. The implementation of assisted migration in Canadian forests. For. Chron. 87, 766–770 (2011).
Allen, M.R. et al. Framing and Context. In: Global Warming of 1.5 °C. IPCC Rep. 51 (IPCC, 2018).
Pedlar, J. H. et al. Placing forestry in the assisted migration debate. Bioscience 62, 835–842 (2012).
Ukrainetz, N. K., O’Neill, G. A. & Jaquish, B. Comparison of fixed and focal point seed transfer systems for reforestation and assisted migration: A case study for interior spruce in British Columbia. Can. J. For. Res. 41, 1452–1464 (2011).
Macdonald, G. B. The case for boreal mixedwood management: an Ontario perspective. For. Chron. 71, 725–734 (1995).
Li, P., Mackay, J. & Bousquet, J. Genetic diversity in Canadian hardwoods: implications for conservation. For. Chron. 68, 709–719 (1992).
Stiell, W. M., Robinson, C. F. & Burgess, D. 20-year growth of white pine following commercial improvement cut in pine mixedwoods. For. Chron. 70, 385–394 (1994).
Ovaskainen, O. et al. Chronicles of nature calendar, a long-term and large-scale multitaxon database on phenology. Sci. Data 7 (2020).
Lesser, M. Genecology, patterns of adaptive variation and a comparison of focal point seed zone development methodologies for white spruce (Picea glauca). (Lakehead University, 2005).
Garrett, P., Schreiner, E., & Kettlewood, H. Geographic variation of eastern white pine in the Northeast. USDA Forest Service Research Paper NE-274 (USDA Forest Service, 1973).
McKenney, D. W. et al. Customized spatial climate models for North America. Bull. Am. Meteorol. Soc. 92, 1611–1622 (2011).
Mackey, B. G., McKenney, D. W., Yang, Y. Q., McMahon, J. P. & Hutchinson, M. F. Site regions revisited: a climatic analysis of Hills’ site regions for the province of Ontario using a parametric method. Can. J. For. Res. 26, 333–354 (1996).
Lawrence, K., Hutchinson, M., & McKenney, D.W. Multi-scale Digital Elevation Models for Canada. Frontline Forestry Research Applications Technical Note No. 109 (Canadian Forestry Service – Sault Ste Marie, 2008).
Risk, C., McKenney, D. W., Pedlar, J. H., & Lu, P. North American tree provenance trial and relevant historical climate data for seven species. The Knowledge Network for Biocomplexity, https://doi.org/10.5063/K072NQ (2020).
O’Neill, G. A., Stoehr, M. & Jaquish, B. Quantifying safe seed transfer distance and impacts of tree breeding on adaptation. For. Ecol. Manage. 328, 122–130 (2014).
Yang, J., Pedlar, J. H., McKenney, D. W. & Weersink, A. The development of universal response functions to facilitate climate-smart regeneration of black spruce and white pine in Ontario, Canada. For. Ecol. Manage. 339, 34–43 (2015).
Schantz-Hansen, T., & Jensen, R. A study of jack pine source of seed. Minnesota Forestry Notes No. 25 (University of Minnesota, 1954).
Shoenike, R., Rudolph, T., & Jensen, R. Branch characteristics in a jack pine seed source plantation. Minnesota Forestry News No. 133 (University of Minnesota, 1962).
Fowler, D. & Heimburger, C. Geographic variation in eastern white pine, 7-year results in Ontario. Silvae Genet. 18, 97–144 (1969).
King, J. & Nienstaedt, H. Variation in eastern white pine seed sources planted in the lake states. Silvae Genet. 18, 83–86 (1969a).
King, J., & Nienstaedt, H. Early growth of eastern white pine seed sources in the lake states. Research Note NC-62 (North Central Forest Experiment Station, USDA Forest Service, 1969b).
Sprackling, J., & Read, R. Jack pine provenance study in eastern Nebraska. USDA Forest Service Research Paper RM-143 (Rocky Mountain Forest and Range Experiment Station, USDA Forest Service, 1975).
Kriebel, H. et al. Geographic variation in Quercus rubra in north central United States plantations. Silvae Genet. 25, 118–122 (1976).
Schlarbaum, S., & Bagley, W. Genetic variation in Quercus rubra L., a Nebraska provenance test report. In Proceedings of the 10th Central States Forest Tree Improvement Conference 157–165 (Department of Forestry & Natural Resources, Purdue University, 1976).
Schlarbaum, S. & Bagley, W. Intraspecific genetic variation of Quercus rubra L., northern red oak. Silvae Genet. 30, 50–56 (1981).
Kriebel, H., Merritt, C. & Stadt, T. Genetics of growth rate in Quercus rubra: provenance and family effects by the early third decade in the north central USA. Silvae Genet. 37, 193–198 (1988).
Chech, F., Keys, R., & Weingartner, D. Seventh-year results of a tamarack provenance study. In Proceedings of the 24th Northeastern Tree Improvement Conference 55–62 (University of Pennsylvania, USDA Forest Service, & Northeastern Forest Experiment Station, 1977).
Chech, F., Keys, R., & Frenchak, B. Fifteen-year results of a limited range eastern larch source study. In Proceedings of the 28th Northeastern Tree Improvement Conference 31–36 (University of New Hampshire & the USDA Forest Service, 1982).
Clausen, K. Within-provenance variation in yellow birch. In Proceedings of the 15th Northeastern Forest Tree Improvement Conference 90–98 (West Virginia University, 1968).
Bihun, Y., & Carter, K. Rangewide provenance study of black spruce in northern Maine (Cooperative Forestry Research Unit Published Article Series No. 31). In Proceedings of the 28th North Eastern Tree Improvement Conference 1–11 (University of New Hampshire & the USDA Forest Service, 1982).
Hale, K. Fourteen year results of a white spruce provenance test planted in Maine. In Proceedings of the 28th North Eastern Tree Improvement Conference 12–15 (University of New Hampshire & the USDA Forest Service, 1982).
Hall, J. Comparison of the growth of larch with other conifers on reforested and afforested sites in Newfoundland. For. Chron. 59, 14–16 (1983).
Park, Y. & Fowler, D. A provenance test of Japanese larch in eastern Canada, including comparative data on European larch and tamarack. Silvae Genet. 32, 96–101 (1983).
Reed, D., Sajdak, R., & Kotar, J. Growth comparisons of tamarack and European larch in Upper Michigan. In Proceedings of the 3rd North Central Tree Improvement Conference 71–77 (USDA Forest Service, 1983).
Khalil, M. All-range black spruce provenance study in Newfoundland: performance and genotypic stability of provenances. Silvae Genet. 33, 63–71 (1984).
Carter, K., & Canavera, D. Jack pine provenance tests in Maine. In Proceedings of the 29th Northeastern Tree Improvement Conference 77–81 (USDA Forest Service & West Virginia University, 1985).
Morgenstern, E., Villeneuve, M., & LeBlanc, R. The range-wide black spruce study: results from 12 experiments in the northeast. In Proceedings of the 30th Northeastern Forest Tree Improvement Conference 188–196 (University of Maine, 1986).
Genys, J. Provenance variation among different populations of Pinus strobus from Canada and the United States. Can. J. For. Res. 17, 228–235 (1987).
Furnier, G., Stine, M., Mohn, C. & Clyde, M. Geographic patterns of variation in allozymes and height growth in white spruce. Can. J. For. Res. 21, 707–712 (1991).
Farmer, R., O’Reilly, G. & Shaotang, D. Genetic variation in juvenile growth of tamarack (Larix laricina) in northwestern Ontario. Can. J. For. Res. 23, 1852–1862 (1993).
van Niejenhuis, A. Genecology of jack pine in north central Ontario. (Lakehead University, 1995).
Morgenstern, K., D’Eon, S. & Penner, M. White spruce growth to age 44 in a provenance test at the Petawawa Research Forest. For. Chron. 82, 572–578 (2006).
Xu, T., & Hutchinson, M. ANUCLIM Version 6.1 User Guide. (Fenner School of Environment and Society, Australian National University, 2010).
Acknowledgements
We would like to recognize the many researchers and field workers who have established, measured, and maintained provenance studies over the years.
Author information
Authors and Affiliations
Contributions
D.M., J.P. and P.L. developed the concept of the study. C.R. collected and entered the data into the two databases, wrote the paper, and created the figures. D.M., J.P. and P.L. reviewed and edited the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online-only Tables
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Risk, C., McKenney, D.W., Pedlar, J. et al. A compilation of North American tree provenance trials and relevant historical climate data for seven species. Sci Data 8, 29 (2021). https://doi.org/10.1038/s41597-021-00820-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-00820-2
This article is cited by
-
Weighted kernels improve multi-environment genomic prediction
Heredity (2023)
-
The Need for a Global Tree Trial Database
New Forests (2023)
-
Study on the variation in and selection of Fraxinus mandshurica provenances and families in northeast China
Journal of Forestry Research (2023)
-
Low genetic variation of foliar traits among Prosopis chilensis (Leguminosae) provenances
Journal of Plant Research (2022)
-
Survival and growth of Acacia senegal (L.) Wild. (Senegalia senegal (L.) Britton) provenances depend on the rainfall at the site of origin
Annals of Forest Science (2021)