Abstract
In lake ecosystems, phytoplankton communities can be studied by adopting taxonomic-based approaches. However, these approaches suffer from identification issues and are sometimes of limited ecosystem ecological value. The recent development of functional approaches may allow an evaluation of other aspects of ecosystem quality, functions and interactions with abiotic parameters or other communities. Here, our aim was to create a phytoplankton trait database at the French scale. This database will be relevant for the analysis of phytoplankton communities that will lead to a better understanding of phytoplankton functional ecology in lakes of France and other European countries possessing similar biological communities. We used a French national database of phytoplankton occurrences sampled from 384 lakes over the entire French metropolitan territory. A final list of 636 taxa was used to compile 53 morpho-functional traits associated with taxonomic information. The traits encompassed variables such as shape, biovolume, motility, toxin production and Reynolds groups. With this new database, we aim to provide data for morpho-functional analyses of phytoplankton assemblages from local to European scale.
Measurement(s) | morphological feature • functional • biovolume • life form • geometric cell form • cell size • carbon content • size • cell motility • mucilage • presence of heterocyte • plastid • presence of siliceous skeleton • cyanotoxin • protuberance • chlorophyll b • chlorophyll c • xanthophyll • nutrition mode • reproduction • trophic water class • Tolerance • Reynolds group • presence of lorica • presence of plate • akinete formation • number of flagella |
Technology Type(s) | digital curation |
Sample Characteristic - Organism | freshwater phytoplankton species |
Sample Characteristic - Environment | lake • freshwater lake |
Sample Characteristic - Location | French River Lakes |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.13416659
Similar content being viewed by others
Background & Summary
In water bodies, phytoplankton communities are key elements of ecosystems, regulating biogeochemical cycles1 and representing one of the most important photoautotrophic primary producer2,3. Because of their rapid responses to water quality and anthropogenic pressures, phytoplankton communities are commonly used as trophic and ecological indicators4,5 in Europe and in the United States6. Therefore, they are mainly studied using a classical taxonomic-based approach, i.e. abundance and biovolume of species. Taxonomic lists are used as a whole, with the sum of individual biovolume employed for total biomass metric assessment7,8, and at the species level for selecting indicator species. This selection is usually based upon the most common species of the phytoplankton community, for which information and ecological profiles can be established9. Therefore, the assessment is only based on a part of the community. Rare species are not considered in such cases although they can be very informative10 (e.g. new invasive species with very specific response to a trophic or a polluted state). Data can be aggregated at higher taxonomic level (family or phylum) to take into account all available taxa7,11. However, different taxa of the same phylum can indicate a very different ecological status while species from different phylum can reflect very similar ecological conditions3. Moreover, the ecological significance can be neutralized at the phylum level by species indicative of opposite conditions12.
During the last decades, approaches based on functional groups were created to overcome these drawbacks13,14,15 and allow evaluating other aspects of ecosystem quality, functioning and biotic interactions16,17. As outlined in Salmaso et al.18 many studies use functional classifications and/or functional groups with the objective to better investigate links between phytoplankton communities and the main environmental factors. Some classifications mentioned by Salmaso et al.18 are based on one characteristic (size classes) or several (morphologically based classifications) while others use functional properties (functional or morpho-functional groups). Indices based on functional groups were also published for lakes19 and for rivers20 to be used for the implementation of Water Framework Directive, WFD21. Nevertheless, as explained by Padisák et al.22 functional groups were not built using all encountered taxa. Ecologists have to adjust their groups manually or through an R code for the Reynolds Functional Groups23. These additions, may not be adequate and lead to misinterpretations. According to Wentzky et al.24, the use of functional-traits at the species level (and not at higher taxonomical level) can limit these issues without influencing any assumption about taxon functional group.
Thus, data collections of phytoplankton traits at the species level are required for a large number of taxa. Accordingly, different datasets were created considering the taxa size. Olenina et al.25 reported size-classes (PEG biovolume, http://ices.dk/data/vocabularies/Documents/Forms/AllItems.aspx) but mainly for marine waters. Kremer et al.26 established a dataset for freshwater and estuaries for a large number of taxa (>1200 species), which had the advantage that it considered cell size and natural unit size (colonial and filamentous growth forms). At the French level, Druart and Rimet created a trait dataset including only biovolumes27 and published, more recently, a phytoplankton trait dataset28 mainly based on Alpine lake surveys and including around 10 compiled traits.
To our knowledge, specific traits such as lorica, protuberance presence (and kind of protuberance), potential toxicity associated with potential toxin production by cyanobacteria, heterocyte and vacuole presence have never been merged together in one trait database. More generally, to our knowledge, no database covers a large number of ecological and morphological traits for phytoplankton communities at the French or European level. To be able to use many morpho-functional traits for water bodies in ecological analyses in France, the construction of such a dataset was initiated. This was based upon the survey database established for the WFD application on French water bodies since 2005. This dataset will be particularly helpful for studying the relationships between functional traits and environmental variables, as well as to develop new trait-based indicators for water quality assessment.
We aim in this paper to (1) compile morpho-functional traits of phytoplankton taxa in French water bodies (metropolitan territory) (2) offer information of the corresponding phylogeny (3) provide a dataset helpful for analyzing functional and morpho-functional phytoplankton assemblages from local to national scale (4) deliver a dataset allowing consistent data comparisons between European countries.
Methods
Site description
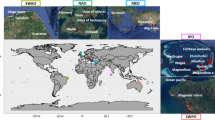
This work was based on the French national database consisting of phytoplankton communities surveyed in 384 lakes between 2005 and 2016 and used for applying the Water Framework Directive. As each lake has been sampled several times this database contains 2,987 phytoplankton samples. Sampled lakes cover the whole French territory (Fig. 1) and encompass all natural and artificial types of lakes encountered in France: from lowlands to alpine location, from small to large lakes (0.09 to 577.12 km2), from shallow to deep lakes (mean depth from 0.30 to 153 meters) and also from oligotrophic to hypereutrophic environmental conditions.
Data acquisition and compilation
In order to create our dataset we firstly defined the taxa list recorded in the national database. The extracted list was firstly examined for taxonomic consistency. Taxa only determined at a high taxonomic level (order, class, phylum) were rejected from the list. Thus, genus and below this level (species, variety, form, etc.) were conserved to obtain an accurate taxonomical list for morpho-functional traits (e.g. biovolume, cell form). A taxa name in the national database can be different over time because of taxonomic changes and determination by different operators. To harmonize taxa denomination and to obtain one up-to-date taxa list, the original names used in the lake database were corrected following the most recent research reported on the AlgaeBase website (https://www.algaebase.org/ - last consultation on 31st July 2020) and in scientific papers. This will allow the traits table to be used for future studies. Thus, a list of 1,318 taxa, associated with abundance and biovolume data, was obtained from the database. In order to remove idiosyncratic species and any potential determination errors, we only kept taxa which occurred in more than 15 samples. A final taxa list of 636 taxa was obtained.
Secondly, we selected a list of 53 morpho-functional traits that we considered to be the most relevant for the description/analyses of functional phytoplankton communities (see Online-only Table 1 for detail of these traits). We compiled available trait data from published sources: we used general books (John et al.29; Reynolds3; Wehr et al.30) for all taxa and specific references listed in Table 1 for more precise information of some traits or taxa. The traits not available in the literature (biovolume, size class, carbon contents, etc.) were determined/calculated. All available synonyms of the taxa recorded in the literature were taken into account in order to consider morpho-functional traits taken from as a many taxonomic references as possible. Taxonomic information was essentially obtained from the AlgaeBase website.
From these publications we mainly obtain morphological characteristics such as cell form, presence of ornamentation on cell and number of chloroplasts. If the information was divergent between two references, the most recent is accepted.
To facilitate the use of the dataset in statistical analyses, taxa code (6 letters in upper case), used in the free computer software Phytobs31 (commonly used in France for freshwater phytoplankton counting), was added for each taxon. These codes were constructed with the first 3 letters for genus and the last 3 for species and infra specific denomination. In order to have a global clustering of phytoplankton taxa (sometimes useful for result presentations), more or less independent of phylogeny updates and thus stable in time, we included a phytoplankton group name. We classified phytoplankton taxa in 9 groups based on common clustering of algae and cyanobacteria such as chlorophytes (green-algae), diatoms, xanthophytes (brown-algae), chrysophytes (gold-algae).
The trait life form was added to the dataset and is defined as the common living form of the taxon in its natural environment (cell, colony or filament). Because some taxa could exhibit a second life form in nature, this information was also indicated in the dataset.
We determined the cell and individual form traits for each taxon with the use of 14 simple geometric forms: sphere, cylinder, rotational ellipsoid, flattened ellipsoid, cymbelloid, staurastrum and ceratium form, oval cylinder, double cone, parallelepiped, prism on parallelogram base and on triangulare base, cone with half sphere and tetrahedron as defined in the computer tool Phytobs31 and illustrated in Fig. 2. Choices were made with the help of book taxa illustrations and according to Hillebrand et al.32, Olenina et al.25, Druart and Rimet27, Hutorowicz33 and Padisak and Adrian34.
Illustration from Phytobs31 of the 14 cell and individual forms used for the biovolume calculations. (a)sphere, (b) cymbelloid, (c) cylinder, (d) oval cylinder, (e) rotational ellipsoid, (f) flattened ellipsoid, (g) ceratium and (h) staurastrum form, (i) double cone, (j) parallelepiped, (k) prism on parallelogram base and on (l) triangulare base, (m) cone with half sphere and (n) tetrahedron.
The minimum and maximum cell dimensions were taken from the literature and were used for the definition of the size class (see below).
We determined taxa biovolume following the European standard NF EN 1669535 with the use of the software Phytobs31 for the calculation (this counting software provides also a biovolume calculation tool). Thus, from real samples observed during the period of the study, 30 individuals of each taxon were measured, as recommended by the European standard, to calculate a mean cell biovolume. For taxa not observed in enough quantity, common minimum and maximum dimensions were taken from the literature in order to determine mean dimensions and calculate a corresponding mean cell biovolume.
We also specified the mean carbon content per cell in our dataset. We used the formulas described in Menden-Deuer and Lessard36 and reproduced in European standard NF EN 1669535 because they consider the decrease in specific carbon content with cell size. Thus, the general formula used for phytoplankton is Carbon [pg C] = 0.216 × Biovolume0.939 and the specific formula for diatom taxa is Carbon [pg C] = 0.288 × Biovolume0.811.
Three size class systems were also subsequently defined in the dataset. The first two use the Sieburth et al.37 class limits and are based on the cell dimensions. The third one uses the biovolume thresholds defined by Ignatiades38. For Sieburth et al. class limits, two variables were defined: one using the minimal individual length and the second using the maximal individual length. In each case, taxa were classified within the following classes: picophytoplankton (0.2–2 µm), nanophytoplankton (2–20 µm), microphytoplankton (20–200 µm), mesophytoplankton (0.2 mm-20 mm). The Ignatiades classification was calculated on individual biovolume: cell biovolume for unicellular taxa and individual biovolume for colonial and filamentous taxa. The classes defined by Ignatiades were: nanoplankton (10–1,000 µm3), microplankton (1,000–1,000,000 µm3), macroplankton (>1,000,000 µm3) plus a specific class, picoplankton (<10 µm3), used for the very small taxa.
Different binary traits (1-presence; 0-absence) were added: motility, presence of flagella, aerotopes, contractile vacuoles, mucilage, akinetes, heterocytes, cysts, plasts, sheath, tractus, siliceous skeleton, lorica, external plates, scales, ornamentations, protuberances, chlorophyll-b and c, xanthophyll, phycobilin and potential toxin production39,40,41,42,43 (with toxin family specified: microcystin, anatoxin-a, anatoxin-a(S), cylindrospermopsin, saxitoxin, Beta-Methylamino-L-Alanin). These characteristics were mainly extracted from the literature. The precise number of plastids and flagella were also added. Some distinctions were defined for the protuberance type, their size and their number onto the individuals. The protuberances were split into seven different types including granule, needle or bristle and the size was divided in two size classes (large and small).
The main nutrition mode was indicated for each taxon following the common definitions: autotroph, mixotroph and heterotroph44,45.
The reproduction mode was specified more precisely when mentioned in the literature or just indicated as sexual or asexual when not.
Some taxa are known only from freshwater and others also from the marine environment. Thus, this information was indicated as a Water_Type in the trait dataset. The trophic status of the water where the taxon lives was defined under “Water_Trophy” as from oligotrophy to hypereutrophy. In the case of a taxon with a large trophic range, the main trophic status was indicated.
In some literature and mainly in Reynolds3, the tolerance to abiotic parameters are defined. We also indicated this trait as a tolerance trait in the dataset (literal form).
We finally reported in the dataset the Reynolds functional groups, based on the original description of Reynolds3 and developed further (with more taxa) by in Borics et al.20 and Padisak et al.19,22.
Data Records
The phytoplankton morpho-functional traits dataset46 is in the form of a large table with taxa in rows and their corresponding morpho-functional traits in columns. This trait table is contained in a formatted file with semicolon-separated values (*.csv) and stored in a public repository available on https://data.inrae.fr/dataverse/eabx. with the following DOI: https://doi.org/10.15454/GJGIAH. The name of the file was defined as FRENCH_PHYTOPLANKTON_TRAITS.csv. This trait table is designed to be updated with new data especially from French lakes (new surveys and new taxa occurrences) and/or new morpho-functional traits relevant for the study of phytoplankton communities.
In the table some variables are numerical and continuous such as biovolumes or cell dimensions, other are categorical such as life forms and other boolean as toxin production. Consequently, the table is a mixed format with numeric and text. The different variables are listed with their attributes in Online-only Table 1 to facilitate understanding and reuse of the data. Missing information was labelled ‘#NA’.
In the dataset, the chlorophytes group is the most represented with 36.5% of the total (Fig. 3) and cryptophytes is the least (2.2%). Other groups like cyanobacteria, diatoms and chrysophytes represent between 10 to 20% of the dataset taxa. The individual life form of each taxon listed is mainly represented by the cell form (56.8%), followed by the colonial form (32.9%) and, finally, the filamentous form of around 10% (Fig. 4).
Technical Validation
The technical validation of the data stored in the dataset was undertaken following the scheme outlined in Fig. 5. All the information collected from the literature was double-checked and all sources are provided in the methods section above. All measures (biovolume, carbon content, size classes attribution, etc.) were made by the same individual in order to reduce error.
Scheme for the technical validation of the dataset. Extraction of the taxa list from the national phytoplankton database, simplification with the synonymy stored in Phytobs software. Implementation of the traits by 3 different operators to obtain a beta version of the dataset, then double-check step and implementation of the calculated traits to obtain a first version of the dataset. Quality control of the data by the referent scientist through the use of graphs and cross-checks to obtain the final version of the dataset.
After that, all data included in the traits table were controlled by the same person (C.L.T.), an expert on French phytoplankton. The dataset thus created was explored through preliminary analyses with box-plots and histograms in order to check distribution and to highlight erroneous data (outliers, aberrant text, missing data, etc.). Many cross-checks were undertaken in order to highlight missing and inconsistent data (C.L.T.):
- motility set to “1” when aerotope, contractile vacuole or flagellum are present;
- number of flagella indicated with presence of flagellum;
- absence of flagellum for desmids, diatoms and cyanobacteria;
- compulsory flagellum for cryptophytes;
- number of plastids indicated with the presence of plastids;
- absence of plastids for cyanobacteria taxa;
- presence of akinete and heterocyte only for cyanobacteria taxa;
- compulsory cyst for chrysophytes and dinophytes taxa;
- siliceous skeleton only for diatoms;
- presence of external plates only for dinophytes;
- presence of lorica for chrysophytes and few chlorophytes and euglenophytes;
- protuberance type other than “None” and protuberance size not null with presence of protuberance;
- presence of chlorophyll-b and xanthophyll and also absence of chlorophyll-c and phycobilin for chlorophytes (equivalent cross-controls for the other phytoplankton groups were done);
- presence of toxin only for some cyanobacteria;
- the toxin family indicated with presence of toxin.
Usage Notes
The phytoplankton trait dataset is freely available and stored in https://data.inrae.fr/dataverse/eabx public repository and should be appropriately referenced by citing the present paper. A survey taxa list should be first checked for synonymy before being analyzed with the morpho-functional dataset. This will avoid the mismatch of taxa names. We strongly encourage users to use the synonymy database present in the Phytobs software.
Note that many traits were obtained from literature indicating general characteristics of species or genera and do not represent individual responses to environmental conditions. Some of them can be affected by different environmental conditions. For example, traits based on dimensions (min, max length and width), biovolume, carbon content and in some cases size-classes can be directly influenced by environmental conditions36,47,48,49 as well as the presence of heterocyte and akinete for some cyanobacteria taxa50.
Code availability
The software Phytobs, used for some trait compilation or calculation (biovolumes), was created by our team in a first version in 2009 by Hadoux and Laplace-Treyture51 for phytoplankton counting purposes. It was upgraded over the years to allow biovolume calculations and to integrate taxonomy and synonymy31. This free software is now in version 3.2 (French, English and Spanish languages) and publicly available on https://hydrobio-dce.inrae.fr/phytobs-software/.
References
Litchman, E. et al. Global biogeochemical impacts of phytoplankton: a trait-based perspective. J. Ecol. 103, 1384–1396 (2015).
De Senerpont Domis, L. N. et al. Plankton dynamics under different climatic conditions in space and time. Freshw. Biol. 58, 463–482 (2013).
Reynolds, C. Ecology of Phytoplankton. (Cambridge University Press, 2006).
Phillips, G. et al. Water Framework Directive Intercalibration: Central Baltic Lake Phytoplankton Ecological Assessment Methods. 189 (Join Research Center, 2014).
Ptacnik, R., Solimini, A. & Brettum, P. Performance of a new phytoplankton composition metric along a eutrophication gradient in Nordic lakes. Hydrobiologia 633, 75–82 (2009).
Pollard, A. I., Hampton, S. E. & Leech, D. M. The promise and potential of continental-scale limnology using the U.S. Environmental Protection Agency’s National Lakes. Assessment. Limnol. Oceanogr. Bull. 27, 36–41 (2018).
de Hoyos, C. et al. Water Framework Directive Intercalibration: Mediterranean Lake Phytoplankton Ecological Assessment Methods. 189 (Join Research Center, 2014).
Mischke, U., Riedmüller, U., Hoehn, E., Schönfelder, I. & Nixdorf, B. Description of the German System for Phytoplankton-Based Assessment of Lakes for Implementation of the EU Water Framework Directive (WFD). 31 (Univ. Cottbus, 2008).
Laplace-Treyture, C. & Feret, T. Performance of the Phytoplankton Index for Lakes (IPLAC): A multimetric phytoplankton index to assess the ecological status of water bodies in France. Ecol. Indic. 69, 686–698 (2016).
Xue, Y. et al. Distinct patterns and processes of abundant and rare eukaryotic plankton communities following a reservoir cyanobacterial bloom. ISME J. 12, 2263–2277 (2018).
Barbe, J. et al. Actualisation de la Méthode de Diagnose Rapide des Plans d’Eau: Analyse Critique des Indices de Qualité des Lacs et Propositions d’Indices de Fonctionnement de l’Écosystème Lacustre. 107 (Cemagref, 2003).
Marchetto, A., Padedda, B., Mariani, M., Luglie, A. & Sechi, N. A numerical index for evaluating phytoplankton response to changes in nutrient levels in deep mediterranean reservoirs. J. Limnol. 68, 106–121 (2009).
Kruk, C., Mazzeo, N., Lacerot, G. & Reynolds, C. S. Classification schemes for phytoplankton: A local validation of a functional approach to the analysis of species temporal replacement. J. Plankton Res. 24, 901–912 (2002).
Reynolds, C. S. Phytoplankton designer – or how to predict compositional responses to trophic-state change. Hydrobiologia 424, 123–132 (2000).
Reynolds, C. S., Huszar, V., Kruk, C., Naselli-Flores, L. & Melo, S. Towards a functional classification of the freshwater phytoplankton. J. Plankton Res. 24, 417–428 (2002).
Mieleitner, J. & Reichert, P. Modelling functional groups of phytoplankton in three lakes of different trophic state. Ecol. Model. 211, 279–291 (2008).
Rangel, L. M., Soares, M. C. S., Paiva, R. & Silva, L. H. S. Morphology-based functional groups as effective indicators of phytoplankton dynamics in a tropical cyanobacteria-dominated transitional river–reservoir system. Ecol. Indic. 64, 217–227 (2016).
Salmaso, N., Naselli-Flores, L. & Padisák, J. Functional classifications and their application in phytoplankton ecology. Freshw. Biol. 60, 603–619 (2015).
Padisák, J., Borics, G., Grigorszky, I. & Soróczki-Pintér, É. Use of phytoplankton assemblages for monitoring ecological status of lakes within the water framework directive: The assemblage index. Hydrobiologia 553, 1–14 (2006).
Borics, G. et al. A new evaluation technique of potamo-plankton for the assessment of the ecological status of rivers. Large Rivers 17, 465–486 (2007).
European Parliament. Directive 2000/60/CE du Parlement Européen et du Conseil du 23 Octobre 2000 Établissant un Cadre pour une Politique Communautaire dans le Domaine de l’Eau. 72 (Communauté Européenne, 2000).
Padisák, J., Crossetti, L. O. & Naselli-Flores, L. Use and misuse in the application of the phytoplankton functional classification: A critical review with updates. Hydrobiologia 621, 1–19 (2009).
Kruk, C. et al. Classification of Reynolds phytoplankton functional groups using individual traits and machine learning techniques. Freshw. Biol. 62, 1681–1692 (2017).
Wentzky, V. C., Tittel, J., Jäger, C. G., Bruggeman, J. & Rinke, K. Seasonal succession of functional traits in phytoplankton communities and their interaction with trophic state. J. Ecol. 108, 1649–1663 (2020).
Olenina, I. et al. Biovolumes and Size-Classes of Phytoplankton in the Baltic Sea. 144 (Baltic Marine Environnment Protection Commission, 2006).
Kremer, C. T., Gillette, J. P., Rudstam, L. G., Brettum, P. & Ptacnik, R. A compendium of cell and natural unit biovolumes for>1200 freshwater phytoplankton species. Ecology 95, 2984–2984 (2014).
Druart, J. C. & Rimet, F. Protocole d’Analyse du Phytoplancton de l’INRA: Prélèvement, Dénombrement et Biovolume. 96 (INRA, 2008).
Rimet, F. & Druart, J.-C. A trait database for phytoplankton of temperate lakes. Ann. Limnol. - Int. J. Limnol. 54, 18 (2018).
John, D. M., Whitton, B. A. & Brook, A. J. The Freshwater Algal Flora of the British Isles: an Identification Guide to Freshwater and Terrestrial Algae. Second Edition. (Cambridge University Press, 2011).
Wehr, J. D., Sheath, R. G. & Kociolek, P. Freshwater Algae of North America: Ecology and Classification. (Academic press, 2015).
Laplace-Treyture, C., Hadoux, E., Plaire, M., Dubertrand, A. & Esmieu, P. PHYTOBS v3.0: Outil de Comptage du Phytoplancton en Laboratoire et de Calcul de l’IPLAC. Version 3.0. Application JAVA. https://hydrobio-dce.inrae.fr/phytobs-software/ (2017).
Hillebrand, H., Dürselen, C. D., Kirschtel, D., Pollinger, U. & Zohary, T. Biovolume calculation for pelagic and benthic microalgae. J. Phycol. 35, 403–424 (1999).
Hutorowicz, A. Opracowanie Standardowych Objętości Komórek do Szacowania Biomasy w Wybranych Taksonów Glonów Planktonowych Wraz z Określeniem Sposobu Pomiarów i Szacowania. 42 (Instytutu Rybactwa Śródlądowego, 2005).
Padisak, J. & Adrian, R. In Methoden der Biologischen Wasseruntersuchung 2. Biologische Gewässeruntersuchung (ed. Friedrich, W. und G.) Biovolumen und Biomasse (Gustav Fischer Verlag, 1999).
NF EN 16695. Qualité de l’eau - Lignes Directrices pour l’Estimation du Biovolume des Microalgues. 106 (2015).
Menden-Deuer, S. & Lessard, E. J. Carbon to volume relationships for dinoflagellates, diatoms, and other protist plankton. Limnol. Oceanogr. 45, 569–579 (2000).
Sieburth, J. M., Smetacek, V. & Lenz, J. Pelagic ecosystem structure: Heterotrophic compartments of the plankton and their relationship to plankton size fractions. Limnol. Oceanogr. 23, 1256–1263 (1978).
Ignatiades, L. Redefinition of cell size classification of phytoplankton – a potential tool for improving the quality and assurance of data interpretation. Mediterr. Mar. Sci. 17, 56 (2015).
Whitton, B. A. Ecology of Cyanobacteria II. Their Diversity in Space and Time. (Springer Verlag, 2012).
Dittmann, E., Gugger, M., Sivonen, K. & Fewer, D. P. Natural product biosynthetic diversity and comparative genomics of the Cyanobacteria. Trends Microbiol. 23, 642–652 (2015).
Sanseverino, I., Conduto, D., Pozzoli, L., Dobricic, S. & Lettieri, T. Algal Bloom and its Economic Impact. 48 (Join Research Center, 2016).
Sanseverino, I., Conduto Antonio, D., Loos, R. & Lettieri, T. Cyanotoxins: Methods and Approaches for their Analysis and Detection. 64 (Join Research Center, 2017).
Meriluoto, J., Spoof, L. & Codd, G. A. Handbook of Cyanobacterial Monitoring and Cyanotoxin Analysis. (John Wiley & Sons, 2017).
Lwoff, A., Van Niel, C. B., Ryan, P. J. & Tatum, E. L. Nomenclature of Nutritional Types of Microorganisms. In Cold Spring Harbor Symposia on Quantitative Biology. XI (5th ed.) 302–303 (1946).
Morris, J. Biology: How Life Works. (W. H. Freeman/Macmillan Learning, 2018).
Laplace-Treyture, C. et al. Phytoplankton morpho-functional trait dataset from French water-bodies. Portail Data INRAE https://doi.org/10.15454/GJGIAH (2020).
Morabito, G., Oggioni, A., Caravati, E. & Panzani, P. Seasonal morphological plasticity of phytoplankton in Lago Maggiore (N. Italy). Hydrobiologia 578, 47–57 (2007).
Naselli-Flores, L., Padisák, J. & Albay, M. Shape and size in phytoplankton ecology: Do they matter? Hydrobiologia 578, 157–161 (2007).
Strathmann, R. R. Estimating the organic carbon content of phytoplankton from cell volume or plasma volume. Limnol. Oceanogr. 12, 411–418 (1967).
Chaffin, J. D., Stanislawczyk, K., Kane, D. D. & Lambrix, M. M. Nutrient addition effects on chlorophyll a, phytoplankton biomass, and heterocyte formation in Lake Erie’s central basin during 2014–2017: Insights into diazotrophic blooms in high nitrogen water. Freshw. Biol. 00, 1–15 (2020).
Hadoux, E. & Laplace-Treyture, C. PHYTOBS: Phytoplankton Counting Tool in Laboratory. Version 1.0. JAVA Application. https://hydrobio-dce.inrae.fr/phytobs-software/ (2009).
Huber Pestalozzi, G. & Thienemann, A. Das Phytoplankton des Susswassers Systematik und Biologie: 5 Teil Chlorophyceae (Grünalgen) Ordnung: Volvocales. (E. Schweizerbart’sche verlagsbuchhandlung, 1974).
Komarek, J., Fott, B. & Huber Pestalozzi, G. Das Phytoplankton des Susswassers Systematik und Biologie: 7 Teil 1 Halfte Chlorophyceae (Grunalgen) Ordnung: Chlorococcales. (E. Schweizerbart’sche verlagsbuchhandlung, 1983).
Coesel, P. F. M. & Meesters, K. J. Desmids of the Lowlands: Mesotaeniaceae and Desmidiaceae of the European Lowlands. (KNNV Publishing, 2007).
Coesel, P. F. M. & Meesters, K. European Flora of the Desmid Genera Staurastrum and Staurodesmus. (KNNV Publishing, 2013).
Starmach, K. Chrysophyceae und Haptophyceae. (VEB Gustav Fischer Verlag, 1985).
Komarek, J. & Anagnostidis, K. Cyanoprokaryota 1.Teil: Chroococcales. (Gustav Fischer, 1999).
Komarek, J. & Anagnostidis, K. Cyanoprokaryota 2.Teil: Oscillatoriales. (Elsevier, 2005).
Komarek, J. Cyanoprokaryota: 3. Teil/Part 3: Heterocytous Genera. (Springer Spektrum Verlag, 2013).
Anses. Evaluation des Risques Liés aux Cyanobactéries et leurs Toxines dans les Eaux Douces. Avis de l’Anses. 438 (Anses, 2020).
Bey, M.-Y. & Ector, L. Atlas des Diatomées des Cours d’Eau de la Région Rhône-Alpes. (DREAL Rhône-Alpes, 2013).
Cox, E. J. Identification of Freshwater Diatoms from Live Material. (Chapman & Hall, 1996).
Druart, J. C. & Straub, F. Description de deux nouvelles Cyclotelles (Bacillariophyceae) de milieux alcalins et eutrophes: Cyclotella costei nov. sp. et Cyclotella wuethrichiana nov. sp. Swiss J. Hydrol. 50, 182–188 (1988).
Houk, V. Atlas of Freshwater Centric Diatoms with a Brief Key and Descriptions Part I Melosiraceae, Orthoseiraceae, Paraliaceae and Aulacoseiraceae. vol. 1 (Czech Phycological Society, Prague & Palacký University Olomouc, 2003).
Houk, V. & Klee, R. Atlas of freshwater centric diatoms with a brief key and descriptions Part II Melosiraceae and Aulacoseiraceae (Supplement to Part I). Fottea J. Czech Phycol. Soc. 7, 85–255 (2007).
Houk, V., Klee, R. & Tanaka, H. Atlas of Freshwater Centric Diatoms with a Brief Key and Descriptions Part IV Stephanodiscaceae B. vol. 14 (Czech Phycological Society, Prague & Palacký University Olomouc, 2014).
Houk, V., Klee, R. & Tanaka, H. Atlas of Freshwater Centric Diatoms with a Brief Key and Descriptions Part III Steogabiduscaceae A Cyclotella, Tertiarius, Discostella. vol. 10 (Czech Phycological Society, Prague & Palacký University Olomouc, 2010).
Houk, V., Klee, R. & Tanaka, H. Atlas of freshwater centric diatoms with a brief key and descriptions: second emended edition of Part I and II Melosiraceae, Orthoseiraceae, Paraliaceae and Aulacoseiraceae. Fottea J. Czech Phycol. Soc. 17, 1–615 (2017).
Krammer, K. & Lange Bertalot, H. Bacillariophyceae. 1. Teil: Naviculaceae. (Specktrum Akademischer Verlag GmbH Heidelberg, 1999).
Krammer, K. & Lange Bertalot, H. Bacillariophyceae. 4. Teil: Achnanthaceae Kritische Ergänzungen zu Achnanthes s.l., Bavicula s. str., Gomphonema. (Spektrum, 2004).
Krammer, K. & Lange Bertalot, H. Bacillariophyceae. 2. Teil: Bacillariaceae, Epithemiaceae, Surirellaceae. (Elsevier, 2007).
Krammer, K. & Lange-Bertalot, H. Bacillariophyceae. 3. Teil: Centrales, Fragilariaceae, Eunotiaceae. (Gustav Fischer Verlag, 2004).
Lange-Bertalot, H., Hofmann, G., Werum, M. & Cantonati, M. Freshwater Benthic Diatoms of Central Europe: Over 800 Common Species Used in Ecological Assessment. (Koeltz Botanical Books, 2017).
Siver, P. A. et al. Observations on Fragilaria longifusiformis comb. nov. et nom. nov. (Bacillariophyceae), a widespread planktic diatom documented from North America and Europe. Phycol. Res. 54, 183–192 (2006).
Popovsky, J. & Pfiester, L. A. Dinophyceae (Dinoflagellida). (Gustav Fischer Verlag, 1990).
Moestrup, Ø. & Calado, A. J. Dinophyceae. vol. 6 (Spektrum Akademischer Verlag, 2018).
Huber Pestalozzi, G. Das Phytoplankton des Susswassers Systematik und Biologie: 4 Teil Euglenophyceen. (E. Schweizerbart’sche verlagsbuchhandlung, 1969).
Ettl, H. Xanthophyceae: 1. Teil. (Gustav Fischer Verlag, 1978).
Rieth, A. Xanthophyceae: 2. Teil. (Gustav Fischer Verlag, 1980).
Acknowledgements
We would like to thank the six water agencies and private labs for participating in the collection of biotic data. We are also grateful to the lake team of Irstea Aix-en-Provence for the lake database management. Many thanks are going to one of our team engineer, S. Boutry, for the map in Fig. 1. We kindly thank the anonymous referees for their useful comments on the previous version of this manuscript. We are also thankful to D.M. John for the English edits. This work was supported by the French Ministry of the ecological and solidarity transition (MTES) and funded by the “Office Français de la Biodiversité” (OFB) through the convention between INRAE and OFB between 2016 and 2018. It was also funded by the Labex COTE “Continental To coastal Ecosystems: evolution, adaptability and governance” within the Cluster of Excellence COTE (ANR-10-LABX-45) and the Adour-Garonne water agency for some personnel costs.
Author information
Authors and Affiliations
Contributions
Christophe Laplace-Treyture (C.L.T.) was the coordinator of the project and the dataset was assembled by Jonathan Derot (J.D.), Anne Le Mat (A.L.M.), Emilie Prévost (E.P.) and Christophe Laplace-Treyture. For the manuscript: conceptualization, methodology, formal illustration and analyses by C.L.T. and A.J.; draft preparation mainly by C.L.T., A.J. with the help of J.D., A.L.M. and E.P. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online-only Table
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Laplace-Treyture, C., Derot, J., Prévost, E. et al. Phytoplankton morpho-functional trait dataset from French water-bodies. Sci Data 8, 40 (2021). https://doi.org/10.1038/s41597-021-00814-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-00814-0
This article is cited by
-
Spatial distribution of sedimentary DNA is taxon-specific and linked to local occurrence at intra-lake scale
Communications Earth & Environment (2023)
-
An integrated individual-level trait-based phytoplankton dataset from transitional waters
Scientific Data (2023)
-
Plankton Community Responses to Anthropogenic Pollution in an Argentinian Urban Reserve
Water, Air, & Soil Pollution (2021)