Abstract
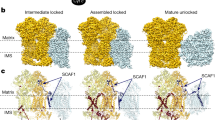
Supercomplexes of the respiratory chain are established constituents of the oxidative phosphorylation system, but their role in mammalian metabolism has been hotly debated. Although recent studies have shown that different tissues/organs are equipped with specific sets of supercomplexes, depending on their metabolic needs, the notion that supercomplexes have a role in the regulation of metabolism has been challenged. However, irrespective of the mechanistic conclusions, the composition of various high molecular weight supercomplexes remains uncertain. Here, using cryogenic electron microscopy, we demonstrate that mammalian (mouse) tissues contain three defined types of ‘respirasome’, supercomplexes made of CI, CIII2 and CIV. The stoichiometry and position of CIV differs in the three respirasomes, of which only one contains the supercomplex-associated factor SCAF1, whose involvement in respirasome formation has long been contended. Our structures confirm that the ‘canonical’ respirasome (the C-respirasome, CICIII2CIV) does not contain SCAF1, which is instead associated to a different respirasome (the CS-respirasome), containing a second copy of CIV. We also identify an alternative respirasome (A-respirasome), with CIV bound to the ‘back’ of CI, instead of the ‘toe’. This structural characterization of mouse mitochondrial supercomplexes allows us to hypothesize a mechanistic basis for their specific role in different metabolic conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Structures of the three SCs were deposited in PDB (accessions 8PW5, 8PW6 and 8PW7) with corresponding cryo-EM density maps in EMDB (IDs 17989, 17990 and 17991). As the models were built on composite maps, the consensus and focused maps for all components of the three SCs have also been deposited on EMDB (18023, 18022, 18025, 18024, 18027, 18026, 18017, 18018, 18019, 18021, 18020, 18015, 18011, 18012, 18013 and 18014). Similarly, structures of complex I were deposited in PDB (accessions 8RGR, 8RGQ, 8RGP and 8RGT) with corresponding composite cryo-EM density maps (19147, 19146, 19145 and 19148) and consensus/focused maps (19085, 19086, 19087, 19091, 19092, 19093, 19088, 19089, 19090, 19105, 19106 and 19107) in EMDB. The following previously deposited models (PDB codes) have been used in the manuscript: 5gup, 5xth, 5iy5, 1ooc, 5z62, 3cx5, 7o3c, 5j4z, 5j7y, 7o37 and 6g2j. Uncropped gels and western blot, as well as the raw data for the activity assays summarized in Extended Data Fig. 8h, have been provided as Source data in this publication. Source data are provided with this paper.
References
Schägger, H. & Pfeiffer, K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 (2000).
Gu, J. et al. The architecture of the mammalian respirasome. Nature 537, 639–643 (2016).
Letts, J. A., Fiedorczuk, K. & Sazanov, L. A. The architecture of respiratory supercomplexes. Nature 537, 644–648 (2016).
Wu, M., Gu, J., Guo, R., Huang, Y. & Yang, M. Structure of mammalian respiratory supercomplex I1III2IV1. Cell 167, 1598–1609.e10 (2016).
Letts, J. A., Fiedorczuk, K., Degliesposti, G., Skehel, M. & Sazanov, L. A. Structures of respiratory supercomplex I+III2 reveal functional and conformational crosstalk. Mol. Cell 75, 1131–1146.e6 (2019).
Vercellino, I. & Sazanov, L. A. Structure and assembly of the mammalian mitochondrial supercomplex CIII2CIV. Nature 598, 364–367 (2021).
Guo, R., Zong, S., Wu, M., Gu, J. & Yang, M. Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell 170, 1247–1257 (2017).
Sousa, J. S., Mills, D. J., Vonck, J. & Kühlbrandt, W. Functional asymmetry and electron flow in the bovine respirasome. eLife 5, e21290 (2016).
Protasoni, M. et al. Respiratory supercomplexes act as a platform for complex III‐mediated maturation of human mitochondrial complexes I and IV. EMBO J. 39, e102817 (2020).
Lobo‐Jarne, T. et al. Multiple pathways coordinate assembly of human mitochondrial complex IV and stabilization of respiratory supercomplexes. EMBO J. 39, e103912 (2020).
Diaz, F., Fukui, H., Garcia, S. & Moraes, C. T. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell. Biol. 26, 4872–4881 (2006).
Schägger, H. et al. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J. Biol. Chem. 279, 36349–36353 (2004).
Ikeda, K. et al. Mitochondrial supercomplex assembly promotes breast and endometrial tumorigenesis by metabolic alterations and enhanced hypoxia tolerance. Nat. Commun. 10, 1–15 (2019).
Wang, G., Popovic, B., Tao, J. & Jiang, A. Overexpression of COX7RP promotes tumor growth and metastasis by inducing ROS production in hepatocellular carcinoma cells. Am. J. Cancer Res 10, 1366–1383 (2020).
Hollinshead, K. E. R. et al. Respiratory supercomplexes promote mitochondrial efficiency and growth in severely hypoxic pancreatic cancer. Cell Rep. 33, 108231 (2020).
Rohlenova, K. et al. Selective disruption of respiratory supercomplexes as a new strategy to suppress Her2high breast cancer. Antioxid. Redox Signal 26, 84–103 (2017).
Antoun, G. et al. Impaired mitochondrial oxidative phosphorylation and supercomplex assembly in rectus abdominis muscle of diabetic obese individuals. Diabetologia 58, 2861–2866 (2015).
Huertas, J. R., Al Fazazi, S., Hidalgo-Gutierrez, A., López, L. C. & Casuso, R. A. Antioxidant effect of exercise: exploring the role of the mitochondrial complex I superassembly. Redox Biol. 13, 477–481 (2017).
Greggio, C. et al. Enhanced respiratory chain supercomplex formation in response to exercise in human skeletal muscle. Cell Metab. 25, 301–311 (2017).
Gonzalez-Franquesa, A. et al. Mass-spectrometry-based proteomics reveals mitochondrial supercomplexome plasticity. Cell Rep. 35, 109180 (2021).
Granata, C. et al. High-intensity training induces non-stoichiometric changes in the mitochondrial proteome of human skeletal muscle without reorganisation of respiratory chain content. Nat. Commun. 12, 7056 (2021).
Frenzel, M., Rommelspacher, H., Sugawa, M. D. & Dencher, N. A. Ageing alters the supramolecular architecture of OxPhos complexes in rat brain cortex. Exp. Gerontol. 45, 563–572 (2010).
Gómez, L. A., Monette, J. S., Chavez, J. D., Maier, C. S. & Hagen, T. M. Supercomplexes of the mitochondrial electron transport chain decline in the aging rat heart. Arch. Biochem. Biophys. 490, 30–35 (2009).
Lombardi, A. et al. Defining the transcriptomic and proteomic profiles of rat ageing skeletal muscle by the use of a cDNA array, 2D- and Blue native-PAGE approach. J. Proteom. 72, 708–721 (2009).
Lopez-Fabuel, I. et al. Complex I assembly into supercomplexes determines differential mitochondrial ROS production in neurons and astrocytes. Proc. Natl Acad. Sci. USA 113, 13063–13068 (2016).
Maranzana, E., Barbero, G., Falasca, A. I., Lenaz, G. & Genova, M. L. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid. Redox Signal 19, 1469–1480 (2013).
Cogliati, S. et al. Mechanism of super-assembly of respiratory complexes III and IV. Nature 539, 579–582 (2016).
Fernández-Vizarra, E. et al. SILAC-based complexome profiling dissects the structural organization of the human respiratory supercomplexes in SCAFI KO cells. Biochim. Biophys. Acta Bioenerg. 1862, 148414 (2021).
Calvo, E. et al. Functional role of respiratory supercomplexes in mice: SCAF1 relevance and segmentation of the Qpool. Sci. Adv. 6, eaba7509 (2020).
García‐Poyatos, C. et al. Scaf1 promotes respiratory supercomplexes and metabolic efficiency in zebrafish. EMBO Rep. 21, e50287 (2020).
Zong, S. et al. Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res 28, 1026–1034 (2018).
Fernández-Vizarra, E. et al. Two independent respiratory chains adapt OXPHOS performance to glycolytic switch. Cell Metab. 34, 1792–1808.e6 (2022).
Benegiamo, G. et al. COX7A2L genetic variants determine cardiorespiratory fitness in mice and human. Nat. Metab. 4, 1336–1351 (2022).
Althoff, T., Mills, D. J., Popot, J.-L. & Kühlbrandt, W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 30, 4652–4664 (2011).
Davies, K. M., Blum, T. B. & Kühlbrandt, W. Conserved in situ arrangement of complex I and III2 in mitochondrial respiratory chain supercomplexes of mammals, yeast, and plants. Proc. Natl Acad. Sci. USA 115, 3024–3029 (2018).
Nesterov, S. et al. Ordered clusters of the complete oxidative phosphorylation system in cardiac mitochondria. Int. J. Mol. Sci. 22, 1–10 (2021).
Letts, J. A. & Sazanov, L. A. Clarifying the supercomplex: the higher-order organization of the mitochondrial electron transport chain. Nat. Struct. Mol. Biol. 24, 800–808 (2017).
Birrell, J. A. & Hirst, J. Truncation of subunit ND2 disrupts the threefold symmetry of the antiporter-like subunits in complex I from higher metazoans. FEBS Lett. 584, 4247–4252 (2010).
Molina-Granada, D. et al. Most mitochondrial dGTP is tightly bound to respiratory complex I through the NDUFA10 subunit. Commun. Biol. 5, 620 (2022).
Vercellino, I. & Sazanov, L. A. The assembly, regulation and function of the mitochondrial respiratory chain. Nat. Rev. Mol. Cell Biol. 23, 141–161 (2021).
Sazanov, L. A. From the ‘black box’ to ‘domino effect’ mechanism: what have we learned from the structures of respiratory complex I. Biochem. J. 480, 319–333 (2023).
Kravchuk, V. et al. A universal coupling mechanism of respiratory complex I. Nature 609, 808–814 (2022).
Kampjut, D. & Sazanov, L. A. The coupling mechanism of mammalian respiratory complex I. Science 370, eabc4209 (2020).
Laube, E., Meier-Credo, J., Langer, J. D. & Kühlbrandt, W. Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum. Sci. Adv. 8, 9952 (2022).
Chung, I. et al. Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy. Nat. Commun. 13, 1–13 (2022).
Agip, A. N. A. et al. Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states. Nat. Struct. Mol. Biol. 25, 548–556 (2018).
Grba, D. N., Chung, I., Bridges, H. R., Agip, A. N. A. & Hirst, J. Investigation of hydrated channels and proton pathways in a high-resolution cryo-EM structure of mammalian complex I. Sci. Adv. 9, eadi1359 (2023).
Gu, J., Liu, T., Guo, R., Zhang, L. & Yang, M. The coupling mechanism of mammalian mitochondrial complex I. Nat. Struct. Mol. Biol. 29, 172–182 (2022).
Dibley, M. G., Ryan, M. T. & Stroud, D. A. A novel isoform of the human mitochondrial complex I subunit NDUFV3. FEBS Lett. 591, 109–117 (2017).
Bridges, H. R., Mohammed, K., Harbour, M. E. & Hirst, J. Subunit NDUFV3 is present in two distinct isoforms in mammalian complex I. Biochim. Biophys. Acta 1858, 197 (2017).
Dyson, H. J. & Wright, P. E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 (2005).
Bianchi, C., Genova, M. L., Castelli, G. P. & Lenaz, G. The mitochondrial respiratory chain is partially organized in a supercomplex assembly: kinetic evidence using flux control analysis. J. Biol. Chem. 279, 36562–36569 (2004).
Lapuente-Brun, E. et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340, 1567–1570 (2013).
Moe, A. et al. Cryo-EM structure and kinetics reveal electron transfer by 2D diffusion of cytochrome c in the yeast III-IV respiratory supercomplex. Proc. Natl Acad. Sci. USA 118, e2021157118 (2021).
Wang, Y. et al. Saturation of the mitochondrial NADH shuttles drives aerobic glycolysis in proliferating cells. Mol. Cell 82, 3270–3283.e9 (2022).
Mühleip, A. et al. Structural basis of mitochondrial membrane bending by the I–II–III2–IV2 supercomplex. Nature 615, 934–938 (2023).
Han, F. et al. Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae. Nat. Commun. 14, 2542 (2023).
Król, S., Fedotovskaya, O., Högbom, M., Ädelroth, P. & Brzezinski, P. Electron and proton transfer in the M. smegmatis III2IV2 supercomplex. Biochim. Biophys. Acta Bioenerg. 1863, 148585 (2022).
Smith, A. L. [13] Preparation, properties, and conditions for assay of mitochondria: slaughterhouse material, small-scale. Methods Enzymol. 10, 81–86 (1967).
Stepanova, A. et al. The dependence of brain mitochondria reactive oxygen species production on oxygen level is linear, except when inhibited by antimycin A. J. Neurochem. 148, 731–745 (2019).
Kun, E., Kirsten, E. & Piper, W. N. Stabilization of mitochondrial functions with digitonin. Methods Enzymol. 55, 115–118 (1979).
Wittig, I., Karas, M. & Schägger, H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteom. 6, 1215–1225 (2007).
Kampjut, D., Steiner, J. & Sazanov, L. A. Cryo-EM grid optimisation for membrane proteins. iScience 24, 102139 (2021).
Zivanov, J., Nakane, T. & Scheres, S. H. W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). 2021 596:7873.
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Acknowledgements
This research was supported by the Scientific Service Units (SSU) of IST Austria through resources provided by the Electron Microscopy Facility (EMF), the Life Science Facility (LSF), the Pre-Clinical Facility (PCF) and the IST high-performance computing cluster. The authors also acknowledge O. Petrova for her help with the complex I data acquisition. I.V. is funded by the ERC Advanced Grant 101020697 RESPICHAIN to L.S. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
I.V. and L.A.S. designed the project. I.V. purified the samples, prepared cryo-EM grids, acquired and processed EM data, built and analyzed the atomic models and wrote the initial draft of the manuscript. L.A.S. acquired funding, supervised the project, analyzed data and models and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors (I.V. and L.A.S.) declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Dimitris Typas was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Representative purification.
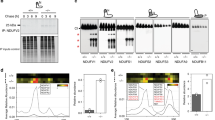
a. Ion exchange chromatography and c. gel filtration profiles with b. and d. respective BN-PAGE of the resulting fractions. (b) Coomassie stain on top and in-gel activity at the bottom, left for CIV and right for CI and (d) Coomassie stain on left, in-gel activity for CI in the middle and for CIV on the right. Peak 3 from the ion exchange was selected and subjected to gel filtration. MW markers are indicated on the left. The experiment was repeated at least three times independently with similar results.
Extended Data Fig. 2 Distribution of supercomplexes across tissues, murine strains and mammalian species.
BN-PAGE runs of CD1 mice hearts (a), brains (b), livers (c), kidneys (d), sheep heart (e) C57 mice hearts (f), brains (g), livers (h), kidneys (i), stained as indicated on top of the figure. SM is solubilised material, P3 is peak 3 of the MonoQ run, WB is Western Blot. The different species are indicated on the right for each section, and the position of 720 kDa MW marker on the left, for clarity. The experiment was repeated at least three times independently with similar results.
Extended Data Fig. 3 Processing of CD1 liver dataset.
Schematic view of the processing pipeline for the CD1 liver dataset, as explained in the Methods section (a), with angular distribution, locally filtered global and focused maps (b), final Fourier Shell Correlation (FSC) graphs for focused maps (c), global maps (d) and models (e).
Extended Data Fig. 4 Processing of CD1 brain dataset and map features.
a. Schematic view of the processing pipeline for the CD1 brain dataset, as explained in the Methods section, with b. final Fourier Shell Correlation (FSC) graphs for the global maps and c. angular distribution and locally filtered global maps. d. Respirasome models derived from the liver dataset fitted into the brain maps. Maps shown in light grey on top by themselves and on bottom with fitted models, coloured as throughout the manuscript and shown as secondary structures. e. Alphafold2 (top left, coloured by confidence values) and Alphafold multimer (top right, four subunits forming NADH-binding domain, coloured by chain) prediction for the structure of the long isoform of NDUFV3, overlaid to the full CI structure (bottom, with full CI in grey). f. Analysis of NDUFV3 long and short isoforms. Left, sequence alignment, where the first 35 residues represent the mitochondrial targeting sequence and the subsequent 25 residues are disordered in short isoform-containing samples, with only ~43 C-terminal residues ordered. Right, western blot of brain CI sample. The experiment was repeated on two independent brain preparation, plus one preparation on different tissues (ovine/murine heart and murine liver) with similar results.
Extended Data Fig. 5 Representative densities from the respirasomes models.
Representative densities for CI (a), CIII2 (b), CIVC (c) and CIVS (d): for each an alpha helix, a beta sheet, a ligand and a lipid are shown. For CI, dGTP is shown in addition to FMN.
Extended Data Fig. 6 CIV conformations and cytochrome c binding in respirasomes.
a-c. Superposition of the structures solved in this manuscript from murine liver (shown as secondary structures with piped helices and coloured as in the other figures, that is CI grey, CIII2 yellow and orange, CIVC cyan with COX7A2 light blue, CIVS ice with SCAF1 dark blue) to the other known mammalian supercomplexes featuring CIV (shown as secondary structures with piped helices and coloured in green). VS is versus. a. Superposition of the murine C-respirasome to the tight (top, PDB 5j4z) and loose (bottom, PDB 5j7y) respirasome from ovine heart, aligned on the membrane arm of CI. The circle highlights the CIV shift between tight and loose conformation. b. Superposition of the CS-respirasome to the tight (top, PDB 5j4z) and loose (bottom, PDB 5j7y) respirasome from ovine heart, aligned on the membrane arm of CI, as in a. As in a, the circles highlight the CIV shift between tight and loose conformation, outlining the region of CIVC clashing between the tight conformation of CIV in the ovine respirasome and the CIVS of the CS-respirasome (top panel). The distance between the edge helices of CI and CIV, depicted by the black lines, is measured in Å in each panel for the ovine (O.a.) and murine (M.m.) supercomplexes shown. In a, the difference in CI-to-CIV distance is 39-31=8 Å between murine C-respirasome and ovine loose respirasome and 39-18=21 Å between murine C-respirasome and ovine tight respirasome. In b, this is 35-31=4 Å between murine CS-respirasome and ovine loose and 35-18=17 Å between murine CS-respirasome and ovine tight. c. Superposition of the CS-respirasome to the unlocked mature (top, PDB 7o3c) and locked (bottom, PDB 7o37) CIII2CIV from murine heart, aligned on SCAF1-containing CIII monomer. The arrows indicate the displacement of CIVS. d-e. Cyt-c binding sites of CS-respirasome. In d, the putative contact between cyt-c bound to CIVS and Lys 46 of COX6B1 on CIVC is depicted: the insets show zoomed-in views of the binding site. In e, the binding sites of cyt-c on CIVS (left) and CIVC (right) are shown. In both panels, cyt-c is docked based on PDB 5iy5, complexes are coloured as in the rest of the manuscript, cyt-c is burgundy.
Extended Data Fig. 7 Interfaces of respirasomes.
Newly-found interaction interfaces in the CS- (a) and A- (b) respirasomes: the details are shown in the insets. CDL is cardiolipin, PC1 is phosphatidylcholine. The complexes are coloured as in the rest of the manuscript, COX6A2 is hot pink and all the lipids are grey.
Extended Data Fig. 8 Complex I structure determination and features.
a-b. Processing overview of CI particles from the liver (a) and brain (b) datasets as described in the Methods section. Representative open (light sea green) and closed (lilac) classes are overlaid in the insets, aligned on the peripheral arm, to highlight the difference in CI conformation between them, used for classification in Focus-Reverse-Classify method. c-e. Final resulting maps, including angular distribution and local resolution (c), half-map FSCs (d) and map-to-model FSCs (e). PA is peripheral arm, MA is membrane arm. f-g. Representative densities for CI from liver (f), and brain (g): as for the other supercomplexes, an alpha helix, a beta sheet, dGTP and FMN ligands and a lipid are shown for closed and open states. For the closed state of liver and brain complex I the quinone density (UQ) is also shown. h-i. CI activity, measured as reduction of A340 absorbance over time due to NADH oxidation. In h, the graph shows the result of three independent purifications from CD1 livers. In the box and whiskers representation, minimum and maximum values are indicated as top and bottom lines; the coloured squares are delimited by first and third quartiles and contain the median value as line with empty dot inside. As the representation results from three independent experiments, the minimum, median and maximum values shown correspond to the individual measurements. No error bars are shown, as no statistical analysis was performed. i shows a representative replicate with raw traces. Active (as prepared) CI is orange, deactive (heated to 37o C for 105 min without substrates, Methods) is grey.
Supplementary information
Source data
Source Data Fig. 1
Unprocessed western blots and gels.
Source Data Extended Data Fig. 1
Unprocessed western blots and gels.
Source Data Extended Data Fig. 2
Unprocessed western blots and gels.
Source Data Extended Data Fig. 4
Unprocessed western blots and gels.
Source Data Extended Data Fig. 8
Raw data and calculated activity for each replicate experiment.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Vercellino, I., Sazanov, L.A. SCAF1 drives the compositional diversity of mammalian respirasomes. Nat Struct Mol Biol (2024). https://doi.org/10.1038/s41594-024-01255-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41594-024-01255-0