Abstract
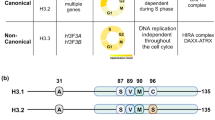
JADE is a core subunit of the HBO1 acetyltransferase complex that regulates developmental and epigenetic programs and promotes gene transcription. Here we describe the mechanism by which JADE facilitates recruitment of the HBO1 complex to chromatin and mediates its enzymatic activity. Structural, genomic and complex assembly in vivo studies show that the PZP (PHD1–zinc-knuckle–PHD2) domain of JADE engages the nucleosome through binding to histone H3 and DNA and is necessary for the association with chromatin targets. Recognition of unmethylated H3K4 by PZP directs enzymatic activity of the complex toward histone H4 acetylation, whereas H3K4 hypermethylation alters histone substrate selectivity. We demonstrate that PZP contributes to leukemogenesis, augmenting transforming activity of the NUP98–JADE2 fusion. Our findings highlight biological consequences and the impact of the intact JADE subunit on genomic recruitment, enzymatic function and pathological activity of the HBO1 complex.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers 8GDX and 8GE0. All other relevant data supporting the key findings of this study are available within the paper, its Supplementary Information or from the corresponding authors upon reasonable request. Source data are provided with this paper.
Code availability
This paper does not report original code.
References
Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 389, 349–352 (1997).
Schubeler, D. et al. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev 14, 940–950 (2000).
Kuo, M. H. & Allis, C. D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20, 615–626 (1998).
Mizzen, C. A. & Allis, C. D. Linking histone acetylation to transcriptional regulation. Cell. Mol. Life Sci. 54, 6–20 (1998).
MacPherson, L. et al. HBO1 is required for the maintenance of leukaemia stem cells. Nature 577, 266–270 (2020).
Hayashi, Y. et al. NUP98-HBO1-fusion generates phenotypically and genetically relevant chronic myelomonocytic leukemia pathogenesis. Blood Adv. 3, 1047–1060 (2019).
Takahashi, S. et al. HBO1-MLL interaction promotes AF4/ENL/P-TEFb-mediated leukemogenesis. eLife 10, e65872 (2021).
Wang, Y. et al. High-expression HBO1 predicts poor prognosis in gastric cancer. Am. J. Clin. Pathol. 152, 517–526 (2019).
Kueh, A. J., Dixon, M. P., Voss, A. K. & Thomas, T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol. Cell. Biol. 31, 845–860 (2011).
Yang, Y. et al. The histone lysine acetyltransferase HBO1 (KAT7) regulates hematopoietic stem cell quiescence and self-renewal. Blood 139, 845–858 (2022).
Miotto, B. & Struhl, K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell 37, 57–66 (2010).
Avvakumov, N. et al. Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation. Mol. Cell. Biol. 32, 689–703 (2012).
Saksouk, N. et al. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol. Cell 33, 257–265 (2009).
Lalonde, M. E. et al. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 27, 2009–2024 (2013).
Han, J. et al. The scaffolding protein JADE1 physically links the acetyltransferase subunit HBO1 with its histone H3-H4 substrate. J. Biol. Chem. 293, 4498–4509 (2018).
Feng, Y. P. et al. BRPF3-HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J. 35, 176–192 (2016).
Peña, P. V. et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442, 100–103 (2006).
Champagne, K. S. et al. The crystal structure of the ING5 PHD finger in complex with an H3K4me3 histone peptide. Proteins 72, 1371–1376 (2008).
Hung, T. et al. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol. Cell 33, 248–256 (2009).
Chen, S. et al. The PZP domain of AF10 Senses unmodified H3K27 to regulate DOT1L-mediated methylation of H3K79. Mol. Cell 60, 319–327 (2015).
Klein, B. J. et al. The role of the PZP domain of AF10 in acute leukemia driven by AF10 translocations. Nat. Commun. 12, 4130 (2021).
Klein, B. J. et al. Molecular basis for the PZP domain of BRPF1 association with chromatin. Structure 28, 105–110 e3 (2020).
Cheng, C. K. et al. A novel NUP98-JADE2 fusion in a patient with acute myeloid leukemia resembling acute promyelocytic leukemia. Blood Adv. 6, 410–415 (2022).
Andrews, F. H., Strahl, B. D. & Kutateladze, T. G. Insights into newly discovered marks and readers of epigenetic information. Nat. Chem. Biol. 12, 662–668 (2016).
Musselman, C. A. & Kutateladze, T. G. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res. 39, 9061–9071 (2011).
Miyamoto, R. & Yokoyama, A. Protocol for fractionation-assisted native ChIP (fanChIP) to capture protein-protein/DNA interactions on chromatin. STAR Protoc. 2, 100404 (2021).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Ruthenburg, A. J. et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145, 692–706 (2011).
Dalvai, M. et al. A scalable genome-editing-based approach for mapping multiprotein complexes in human cells. Cell Rep. 13, 621–633 (2015).
Altaf, M. et al. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 28, 1002–1014 (2007).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Klein, B. J. et al. The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep. 6, 325–335 (2014).
Gatchalian, J. et al. Dido3 PHD modulates cell differentiation and division. Cell Rep. 4, 148–158 (2013).
Becht, D. C. et al. MORF and MOZ acetyltransferases target unmethylated CpG islands through the winged helix domain. Nat. Commun. 14, 697 (2023).
Tencer, A. H. et al. Covalent modifications of histone H3K9 promote binding of CHD3. Cell Rep. 21, 455–466 (2017).
Musselman, C. A. et al. Binding of PHF1 tudor to H3K36me3 enhances nucleosome accessibility. Nat. Commun. 4, 2969 (2013).
Yokoyama, A., Kawaguchi, Y., Kitabayashi, I., Ohki, M. & Hirai, K. The conserved domain CR2 of Epstein-Barr virus nuclear antigen leader protein is responsible not only for nuclear matrix association but also for nuclear localization. Virology 279, 401–413 (2001).
Yokoyama, A. et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 24, 5639–5649 (2004).
Okuda, H. & Yokoyama, A. Myeloid progenitor transformation assay. Bio Protoc. 7, e2626 (2017).
Acknowledgements
We thank H. Sato, I. Yokoyama, K. Ito, E. Kanai and A. Yokoyama for technical assistance. This work was supported in part by grants from the National Institutes of Health: HL151334, GM135671, GM125195, CA252707 and AG067664 to T.G.K., GM131626 and GM139564 to M.G.P., from the Japan Society for the Promotion of Science (JSPS) KAKENHI grants (22H03109 and 22KK0119) to A.Y. and A.K. and from the Canadian Institutes of Health Research (CIHR) (FDN-143314, PJT-178367) to J.C. This work was also supported in part by research funds to A.Y. from the Yamagata prefectural government and the city of Tsuruoka.
Author information
Authors and Affiliations
Contributions
N.G., A.K., C.L., K.L.C., J.L., A.T.G., N.S., B.J.K., Y.K. and S.A. performed experiments and, together with A.J.R., M.G.P., J.C., A.Y. and T.G.K., analyzed the data. A.Y. and T.G.K. wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Jun Qin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editors: Sara Osman and Carolina Perdigoto, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and Tables 1–3.
Supplementary Data
Source data for Supplementary Information.
Source data
Source Data Fig. 7
ChIP–seq tags, ChIP–qPCR, expression and colony-forming data.
Source Data Figs. 1, 2 and 6
Uncropped blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gaurav, N., Kanai, A., Lachance, C. et al. Guiding the HBO1 complex function through the JADE subunit. Nat Struct Mol Biol (2024). https://doi.org/10.1038/s41594-024-01245-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41594-024-01245-2