Abstract
Integrin affinity regulation, also termed integrin activation, is essential for metazoan life. Although talin and kindlin binding to the β-integrin cytoplasmic tail is indispensable for integrin activation, it is unknown how they achieve this function. By combining NMR, biochemistry and cell biology techniques, we found that talin and kindlin binding to the β-tail can induce a conformational change that increases talin affinity and decreases kindlin affinity toward it. We also discovered that this asymmetric affinity regulation is accompanied by a direct interaction between talin and kindlin, which promotes simultaneous binding of talin and kindlin to β-tails. Disrupting allosteric communication between the β-tail-binding sites of talin and kindlin or their direct interaction in cells severely compromised integrin functions. These data show how talin and kindlin cooperate to generate a small but critical population of ternary talin–β-integrin–kindlin complexes with high talin–integrin affinity and high dynamics.
Similar content being viewed by others
Main
Integrins adhere cells to the extracellular matrix, probe biochemical and biophysical properties of the extracellular matrix and convert the information into cellular responses such as spreading, migration, proliferation, survival and differentiation.
Integrins are α–β heterodimers consisting of a large ectodomain, a single-span transmembrane (TM) helix and a short C-terminal cytoplasmic tail (CT). A hallmark of integrins is the ability to reversibly switch between conformations with low and high affinity for ligand. The affinity switch requires the binding of the adaptor proteins talin and kindlin to the β-integrin CT (β-CT). In mammals, there are two talin (TLN1 and TLN2) and three kindlin (KIND1, KIND2 and KIND3) isoforms. Talin and kindlin colocalize in integrin-containing focal adhesions and cooperate to enable integrin–ligand binding and cell adhesion1,2,3. The mechanism underlying this cooperation remains a major unresolved question in adhesion biology.
Talin and kindlin are FERM (protein 4.1, ezrin, radixin, moesin) domain proteins consisting of F1, F2 and F3 subdomains and an additional N-terminal talin- and kindlin-specific ubiquitin-like F0 subdomain4,5. The F2 subdomain of kindlin harbors a pleckstrin homology (PH) domain, which binds negatively charged lipids such as phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2 or PIP2) and phosphatidylinositol (3,4,5)-trisphosphate (PtdIns(3,4,5)P3 or PIP3)6,7,8. The talin FERM domain, called the talin head domain (THD), binds charged membrane lipids such as PIP2 (ref. 9) and is connected via a flexible linker peptide to an elongated, mechanosensitive F-actin-binding rod domain10. The F3 subdomains of talin and kindlin fold into a phosphotyrosine-binding domain and bind distinct and juxtaposed regions in β-CTs. NMR and crystallographic studies show that talin F3 binds the membrane-proximal NPxY motif and the α-helical region of the β3-CT11,12, and kindlin F3 binds the membrane-distal NxxY motif4,13.
A ternary talin–β-integrin–kindlin complex has been observed for different kindlin and β-integrin isoforms by NMR spectroscopy, analytical ultracentrifugation or super-resolution microscopy14,15,16 and is considered crucial for talin–kindlin cooperativity. However, this hypothesis has not been proven so far. Theoretically, one would expect that talin and kindlin binding to the β-CT cooperate by amplifying each other’s activity10. In the case of the β3-CT, surface plasmon resonance experiments point to independent binding of TLN1 and KIND2 to the β3-CT14. On the contrary, pulldown experiments reported competition of recombinant KIND2 and KIND3 by TLN1 (ref. 17), and molecular dynamics simulations suggested that talin and kindlin differentially influence each other during β3-CT binding18.
Here, we investigated the mechanistic basis of talin and kindlin association with β-CT. We found that ternary interactions of THD and KIND2 with β1-CT or β3-CT can induce an allosteric change of the β-CT, which increases THD and decreases KIND2 affinity. We also observed a direct talin–kindlin interaction, which likely enables rebinding and maintaining the populations of ternary talin–β-integrin–kindlin complexes at a critical threshold. The complex cooperativity between talin and kindlin results in an intrinsic cycle of assembly and disassembly of ternary talin–β-integrin–kindlin complexes that is solely governed by molecular communication between talin and kindlin.
Results
β1-tail, TLN1 and KIND2 form a ternary complex at equilibrium
To determine whether the β1A CT splice isoform (β1-CT) assembles, similar to β3-CT14, a ternary complex with TLN1 and KIND2 in vitro, we used NMR spectroscopy to characterize the interaction of isotope-labeled β1-CT with the unlabeled F3 domain of TLN1 (TLN1-F3) and KIND2 lacking the flexible loop in F1 and the PH domain (ΔKIND2; Fig. 1a and Extended Data Fig. 1a) to reduce molecular weight and enhance solubility and NMR spectral quality. NMR [1H,15N and 1H,13C]methyl correlation spectra (Fig. 1b-d and Extended Data Fig. 1b,c) are affected by the presence of a binding partner, and changes appear either as chemical shift perturbation (CSP), defined as shift of an NMR signal (Extended Data Fig. 1b), or line broadening leading to decreasing signal intensity (Fig. 1b and Extended Data Fig. 1e). Interestingly, ΔKIND2 binding affected the β1-CTG778 residue, which is located in the talin-binding site, whereas TLN1-F3 binding did not affect β1-CTG797 in the kindlin-binding site, pointing to a different mutual influence of kindlin and talin during β1-CT binding. Of note, the line width of an NMR signal is related to the molecular weight of the protein tumbling in solution. Thus, the line broadening observed during complex formation will reflect the increase in the molecular weight of the complex. In addition, the stability of the complex, that is, the binding off-rate and local conformational dynamics due to interaction with the binding partners, can provide an additional contribution to the line width. Residues in β1-CT undergoing the greatest line broadening shown by NMR signal are those in the talin- or kindlin-binding sites. The extent of line broadening likely reflects the molecular weight of the complex, although additional contributions from binding kinetics or conformational dynamics cannot be excluded.
a, Proteins used in this study. Amino acid numbering of isoforms is shown in brackets. ΔKIND2 lacks the flexible loop (star; Δ168–217) and the PH domain (Δ337–512). b, Overlay of one-dimensional traces of amide signals of residues G778 and G797 in talin- and kindlin-binding regions of β1-CT in 1H–15N HSQC NMR spectra of 15N-labeled β1-CT before (black) and after addition of increasing stoichiometries of ΔKIND2 (top, green) or TLN1-F3 (middle, blue) or both TLN1-F3 and ΔKIND2 (bottom, purple). AU, arbitrary units. c, Intensity ratio of peaks of β1-CT in the presence and the absence of ΔKIND2 (top, green), TLN1-F3 (middle, blue) or both (bottom, magenta). Talin- (blue) and kindlin- (green) binding sites are indicated above plots (MP, membrane proximal; MD, membrane distal). The isolated peaks in b are indicated by (1) amino acid numbering, shown below plots, (2) dashed lines and (3) reported mean intensity values for each titration in their respective color code. d, Magnified view of [1H,13C]methyl correlations observed for 100 µM 13C,15N-labeled β1-CT before (black) and after addition of ΔKIND2 (green) or TLN1-F3 (blue) or TLN1-F3 and ΔKIND2 (purple). Methyl groups of A764 (α-helical), A773 (membrane-proximal NPxY) and A786 (membrane-distal NPxY) are shown. Arrows indicate chemical shifts induced by protein addition. e, MST measurements of THD1 affinity for 488–β3-CT in the presence (orange) or the absence (blue) of 30–60 µM KIND2 (3–6-fold excess of Kd). f, MST measurements of KIND2 affinity for 488–β3-CT in the presence (purple) or the absence (green) of 250–450 µM (2–4-fold excess of Kd) THD1 or 100–400 µM (2–10-fold excess of Kd) THD2 (purple). g, Binding affinities of THD1 and/or KIND2 for β3-CT are inconsistent with the simple ternary-complex model. h, Exemplary dose–response curves from FC-RDA showing competition of ATTO 565-labeled THD1 (565–THD1) with unlabeled THD1 (blue) or unlabeled KIND2 (magenta) and competition of Alexa 647-labeled KIND2 (647–KIND2) with unlabeled KIND2 (green) or unlabeled THD1 (orange) for β1-TM–CT embedded in PIP2-containing nanodiscs. Mean fluorescence intensity (MFI) was normalized to positive and negative values. Dashed lines indicate the IC50 value of 565–THD1 competing with unlabeled THD1 (blue) or unlabeled KIND2 (purple). i, FC-RDA-generated IC50 values of unlabeled THD1 competing with 565–THD1 (blue) and 647–KIND2 (orange) and unlabeled KIND2 competing with 647–KIND2 (green) and 565–THD1 (purple) for β1-TM–CT, β1-TM–CTY795A, β1-TM–CTY783A and α5-TM embedded in PIP3-containing nanodiscs and β3-TM–CT embedded in PIP2- or PIP3-containing nanodiscs. Bars display mean ± s.d. with a line for the median.
Superposition of 1H–15N correlation spectra of β1-CT in the absence and the presence of TLN1-F3 or ΔKIND2 shows reduced peak intensities assigned to a region from A773 to A786 including the membrane-proximal NPxY motif for TLN1-F3 and from Y783 to the C-terminal end of the β1-CT including the membrane-distal NPxY motif for ΔKIND2 (Fig. 1b,c and Extended Data Fig. 1e). This finding agrees well with the reported talin- and kindlin-binding sites4,11,12. Furthermore, 1H–15N (Fig. 1c and Extended Data Fig. 1e) and 1H–13C (Fig. 1d and Extended Data Fig. 1c) correlation spectra show minor CSPs and small changes in signal intensities assigned to the membrane-proximal, α-helical region from H758 to K770, suggesting that, in contrast to β3-CT, this conserved region is not an important binding site for TLN1-F3 in β1-CT12,19. Interestingly, spectral peak intensities assigned to the β1-CTY783–A786 region are reduced upon addition of TLN1-F3 as well as ΔKIND2, suggesting that talin- and kindlin-binding sites overlap in β1-CT (Fig. 1c).
To test whether talin and kindlin form a ternary complex with β1-CT, we titrated both ΔKIND2 and TLN1-F3 to isotope-labeled β1-CT. While the chemical shifts in the α-helical region (H758–K770) remained almost unchanged (Extended Data Fig. 1b), the average signal intensities throughout the entire β1-CT decreased in the presence of increasing ΔKIND2 and TLN1-F3 concentrations compared to spectra of binary TLN1-F3–β1-CT or ΔKIND2–β1-CT complexes (Fig. 1c and Extended Data Fig. 1e). This global signal intensity decrease in the β1-CT is consistent with a molecular weight increase upon formation of a ternary complex. In addition, the 1H–13C correlation spectra of β1-CT in the presence of both TLN1-F3 and ΔKIND2 (Fig. 1d and Extended Data Fig. 1c) reveal that methyl signals of several amino acids between the membrane-proximal and -distal NPxY motifs (A786–V791) are line broadened beyond detection, which is neither observed for all the other peaks nor is it in binary β1-CT–TLN1-F3 or β1-CT–ΔKIND2 complexes. This observation indicates changes in the binding regime (on and off kinetics) during formation of the ternary TLN1-F3–β1-CT–ΔKIND2 complex and possibly a conformational change of β1-CT upon simultaneous binding of TLN1-F3 and ΔKIND2. Because simultaneous as well as sequential addition of TLN1-F3 and ΔKIND2 to labeled β1-CT produces identical 1H–13C correlation spectra, we conclude that the ternary complex forms at thermodynamic equilibrium (Extended Data Fig. 1d).
Talin and kindlin affinities for β-tails
To investigate whether talin and kindlin influence each other upon β1-CT or β3-CT binding, we determined their dissociation constants (Kd values) for β-CTs at thermodynamic equilibrium. In the simplest case of ternary talin–β-CT–kindlin complex formation, the assumption is that either talin or kindlin binds to β-CT first and then the remaining free adaptor binds to the occupied β-CT, which can be illustrated with an ‘energy square’ (equation (1)):

Measurement of the dissociation constant of talin or kindlin for the β-CT that is either free (Kd) or occupied with kindlin or talin (Kd*) allows us to prove or disprove the ternary-complex model. As the total energy does not change in a closed system, microscopic reversibility demands that the product of dissociation and association constants (Ka = 1/Kd) around a reaction cycle of an energy square must equal 1 (equation (2):
Furthermore, a comparison of the dissociation constants of talin and kindlin for free or occupied β-CT allows us to differentiate independent (Kd = Kd*), competitive (Kd < Kd*) or mutually reinforced binding of talin and kindlin to β-CT (Kd > Kd*) and assign a potential function to the ternary talin–β-integrin–kindlin complex.
We determined the talin- and kindlin-binding mode to β1-CT by microscale thermophoresis (MST)20, which quantifies changes in fluorescence induced by a temperature-related intensity change as well as thermophoresis of a fluorescently labeled probe. The extent of temperature-related intensity change due to ligand binding and thermophoresis due to size, charge and solvation entropy differences were used to quantify binding affinities in titration experiments. To minimize heat effects, we measured the fluorescence changes only 1.5 s before and after turning on the infrared laser (Extended Data Fig. 1f). MST-based Kd measurements of recombinant THD1, THD2 and KIND2 (Fig. 1a and Extended Data Fig. 1a) for ATTO 488-labeled (488)–β1-CTs were performed in the absence and the presence of KIND2, THD1 or THD2 at near-saturation binding concentrations (Extended Data Fig. 1f–h). The maximal solubilities of KIND2 and THD1, which were about 500 µM (40 mg ml−1) and 2 mM (100 mg ml−1), respectively, allowed only near-saturation binding experiments and the determination of apparent affinities for β-CTs.
MST experiments revealed that near-saturation binding levels of KIND2 did not interfere with THD1 binding to 488–β1-CT, resulting in Kd,THD1 = Kd,app,THD1* (equations (1) and (2)). Unexpectedly, THD1 at near-saturation binding concentrations decreased KIND2 binding to β1-CT (Extended Data Fig. 1g,h), resulting in Kd,KIND2 < Kd,app,KIND2*. However, measurements with the 488–β1-CT produced bell-shaped binding curves in the presence of KIND2 (Extended Data Fig. 1h), which are quite frequently observed in MST traces caused by unknown physical phenomena21. To obtain more accurate MST measurements, we measured binding to 488–β3-CTs, which produced sigmoid, one-site-binding curves when plotted against increasing concentrations of THD1, THD2 and KIND2, respectively, with dissociation constants of 108 ± 12 µM for THD1, 39 ± 3 µM for THD2 and 9 ± 2 µM for KIND2 (Fig. 1e,f, Extended Data Fig. 1i and Extended Data Table 1). In agreement with β1-CT competition measurements (Extended Data Fig. 1g,h), the apparent dissociation constants of THD1 or THD2 for β3-CT were unaffected by the presence of KIND2 (Kd,THD = Kd,app,THD*; Fig. 1e and Extended Data Fig. 1i), whereas the apparent dissociation constants of KIND2 increased to 54 ± 27 µM, 120 ± 1 µM, 27 ± 5 µM and 47 ± 14 µM in the presence of THD1, TLN1-F3, THD2 and TLN2-F3, respectively (Kd,KIND2 < Kd,app,KIND2*; Fig. 1f and Extended Data Fig. 1j). Strikingly, insertion of MST-derived dissociation constants of THD1, KIND2 and β3-CT in the energy square did not equal 1 but produced a value of 0.16 ± 0.11:
The unidirectional competitive behavior of THD1 and KIND2 for β3-CT (and β1-CT) at chemical equilibrium contradicts a simple ternary-complex model (Fig. 1g and equation (1)) and suggests that binding of talin and kindlin to β-CT is much more complex.
To confirm unidirectional competition between talin and kindlin with an orthogonal assay that includes lipid-binding sites for THD1 and KIND2, we incorporated recombinant, biotinylated β1 and β3 TM- and CT-containing polypeptides (β1-TM–CT, β3-TM–CT; Fig. 1a) into 10% phosphatidylinositol phosphate- and 90% phosphocholine-containing nanodiscs (Extended Data Fig. 1a). The reconstituted nanodiscs were immobilized on streptavidin beads and analyzed in a flow cytometry-based reporter-displacement assay (FC-RDA) that allowed us to determine the concentration of unlabeled THD1 or KIND2 required to decrease binding of fluorescently labeled THD1 and KIND2 to 50%, respectively (IC50; Fig. 1h,i). The IC50 values of unlabeled THD1 competing with fluorescently labeled THD1 and of unlabeled KIND2 competing with fluorescently labeled KIND2 report affinities. The IC50 values of unlabeled THD1 competing with fluorescently labeled KIND2 and of unlabeled KIND2 competing with fluorescently labeled THD1 report the capability to displace the other adaptor, which is related to the apparent dissociation constant Kd,app*, measured by MST. FC-RDA measurements revealed that affinities of THD1 as well as KIND2 for β1-TM–CTs and β3-TM–CTs embedded in PIP2- or PIP3-containing nanodiscs were in the range of around 80–100 nM and, for α5-TM lacking the cytoplasmic domain (tailless α5-TM) embedded in PIP2- or PIP3-containing nanodiscs, were in the range of around 300–500 nM. As the tailless α5-TM interacts with neither talin nor kindlin, these findings indicate that charged lipids contribute the largest binding energy for talin and kindlin, whereas β-tails make a minor contribution (Extended Data Table 1), which is in line with reports for THD1 and β3-CT22. The ability of THD1 to compete with labeled KIND2 and of KIND2 with labeled THD1 with similar IC50 values from tailless α5-TM embedded in PIP2- or PIP3-containing nanodiscs indicates that talin and kindlin compete with similar efficiency for the same lipid-binding sites. In line with the data obtained by MST (Fig. 1g), KIND2 was unable to effectively outcompete THD1 binding to β1-TM–CTs as well as β3-TM–CTs embedded in PIP3- or PIP2-containing nanodiscs, whereas THD1 readily displaced KIND2 (Fig. 1i and Extended Data Fig. 1k). MST data also showed that THD1 failed to displace KIND2 from talin-binding-impaired β1-CTY783A and β3-CTY772A (Extended Data Fig. 1l,m), which altogether indicates that THD1 displaced KIND2 in a β-CT-binding-dependent manner.
KIND2 directly binds TLN1 to stabilize the ternary complex
To achieve microscopic reversibility, the decrease in KIND2 affinity for β3-CT in the presence of THD1 (Fig. 1g) must be accompanied by the same decrease in THD1 affinity in the presence of KIND2. Because THD1 affinity, however, does not decrease in the presence of KIND2, we hypothesized that allostery in the β-CT and/or between talin and KIND2 caused by a direct talin–KIND2 interaction counteracts the decrease in THD1 affinity and may lead to unidirectional binding preference. To investigate the talin–kindlin interaction, we probed for an interaction of TLN1-F3 with KIND2 in the absence and presence of β1-CT using NMR. First, we examined whether TLN1-F3 and KIND2 bind in the absence of β-CTs by adding KIND2 in a 0.75-fold ratio to uniformly 15N-labeled TLN1-F3. Analysis of 1H–15N heteronuclear single quantum coherence (HSQC) NMR spectra revealed CSPs in the α1-helix of TLN1-F3 located between the integrin-binding site and the flexible linker segment that joins THD and the rod domain in full-length talin (Fig. 2a–c and Extended Data Fig. 2a). NMR spectral changes in the α1-helix of TLN1-F3 differ from those observed in NMR spectra of binary complexes between β1-CT and uniformly 15N-labeled TLN1-F3 (Extended Data Fig. 2b–d), indicating that the KIND2 and β-CT-binding sites localize to adjacent but distinct regions of the TLN1-F3 domain. This finding was confirmed by adding KIND2 to uniformly 15N-labeled TLN1-F3 in the presence of β1-CTs at saturating concentrations, which induced CSPs in the α1-helix as well as in the integrin-binding site of TLN1-F3 (Extended Data Fig. 2e–g).
a,b, Overlay of 1H–15N HSQC NMR spectra (a) and CSP plot (b) of 15N-labeled TLN1-F3 in the absence (black) and the presence (green) of KIND2. c, Significant CSP changes (>0.018 ppm) are indicated by the dashed line (2σ interval, P < 0.05) and mapped on the TLN2-F3 crystal structure (PDB 3G9W). Amino acid numbering is below the plots; TLN1K402 is shown in purple. d, Overlay of 1H–15N HSQC NMR spectra of 70 µM 15N-labeled wild-type TLN1-F3 (top) and TLN1-F3K402E (bottom) in the absence (black) and the presence of 140 µM (dark green) and 700 µM (light green) ΔKIND2. Most peaks in TLN1-F3 spectra disappear (top) and remain in TLN1-F3K402E upon ΔKIND2 titration (bottom). Note that contour line thickness is adjusted in both images to improve visibility. e, Model of the TLN-F3–β1-CT–ΔKIND2 complex based on chemical cross-links identified by mass spectrometry (Extended Data Fig. 2I) and reported structures of TLN2-F2F3 (blue)–β1D-CT (gray; PDB 39GW), ΔKIND2 (green)–β1A-CT (gray; PDB 5XQ0) and KIND2-F0 (green; PDB 2LGX). Cross-links between relevant distance of 5–30 Å are colored in yellow; further cross-links are in red. f, Model indicating residues in KIND2 that were analyzed further. g, Affinity reduction of THD1 for 488–β3-CT in the absence (Kd,THD1) and the presence of 30–40 µM KIND2 (Kd,THD1+KIND2) with indicated substitutions in KIND2 (n = 2), measured by MST. h,i, Magnified view of an overlay of 1H–15N HSQC NMR spectra of 100 µM 15N-labeled KIND2-F0 in the absence (black) and the presence of 100 µM TLN1-F3 (blue) or 100 µM TLN1-F3K402E (red) (h) and 1H–15N HSQC NMR spectra of 70 µM 15N-labeled KIND2-F0Y13A in the absence (black) and the presence (purple) of 70 µM wild-type TLN1-F3 (i). j, Ratio of average mean peak intensities (±s.d.) assigned to residues H758–K770 in the α-helical region of 15N-labeled β1-CT at 1:0.8 and 1:0.2 ratios of TLN1-F3 and/or ΔKIND2 (raw data in Fig. 1c and Extended Data Fig. 2l).
To narrow down the KIND2-binding site, we substituted each amino acid in the TLN1-F3 α1-helix individually with alanine or glutamic acid to identify THD1 mutants that affect affinities for β3-CT in the presence of near-saturation binding concentrations of KIND2. MST measurements revealed that, among all THD1 mutants tested (Extended Data Fig. 2h), only K401, K402 or K403, double substitutions of KK402/403 or deletions of amino acids 401–405 in THD1 lowered the affinity for β3-CTs in the presence of KIND2. A comparison of NMR spectra of 15N-labeled TLN1-F3 and TLN1-F3K402E in the presence of ΔKIND2 confirmed the involvement of TLN1-F3K402 for KIND2 binding. Almost all NMR signals in the wild-type TLN1-F3 spectra experience severe line broadening beyond detection in the presence of ΔKIND2 at a molar excess of 2–10-fold. Under these conditions, the substantial molecular weight increase of the TLN1-F3–ΔKIND2 complex (from 11.6 kDa to 65.1 kDa) and potential dynamics in the binding interface led to substantial line broadening beyond detection. However, NMR signals in the TLN1-F3K402E spectra remain largely unaffected upon ΔKIND2 titration, consistent with the substantially reduced binding of TLN1-F3K402E to ΔKIND2 (Fig. 2d).
To define the corresponding talin-binding site on KIND2, we fused the C terminus of TLN1-F3 via a flexible glycine–serine (GS) linker to the N terminus of β1-CT to produce recombinant TLN1-F3–β1-CT or TLN1-F3K402E–β1-CT fusion proteins with increased talin–integrin binding events. The fusion proteins were cross-linked with disuccinimidyl suberate in the presence of excess KIND2, and cross-links were identified by mass spectrometry. We found numerous cross-links between TLN1-F3 and the F0 domain of KIND2 (KIND2-F0) and very few between TLN1-F3K402E and KIND2-F0 (Extended Data Fig. 2i). Next, we used the cross-links to build a model of a ternary TLN2-F3–β1-CT–ΔKIND2 complex derived from published crystal structures (Fig. 2e) and noticed that the flexible N-terminal loop of KIND2 may be close to the α1-helix of TLN1-F3 (Fig. 2f). As four lysine residues in the TLN1-F3 α1-helix comprise the binding site for KIND2-F0 (Extended Data Fig. 2h), we expected either negatively charged or aromatic residues on the KIND2-F0 countersurface as the contact site (Fig. 2f). Substitution of the positively charged and aromatic residues in the flexible N terminus of the F0 domain of recombinant KIND2 for lysine and alanine, respectively, revealed that only KIND2Y13A decreased THD1 affinity for 488–β3-CT in MST measurements (Fig. 2g). To confirm the interaction between TLN1K402 and KIND2Y13, we titrated TLN1-F3 or TLN1-F3K402E with the 15N-labeled KIND2-F0 domain, recorded 1H–15N-HSQC NMR spectra and observed that TLN1-F3 induces stronger CSPs in the KIND2-F0 domain than TLN1-F3K402E (Fig. 2h and Extended Data Fig. 2j–m). Furthermore, almost all CSPs induced by TLN1-F3 titration with 15N-labeled KIND2-F0 drastically decreased when TLN1-F3 was titrated with 15N-labeled KIND2-F0Y13A (Fig. 2i and Extended Data Fig. 2k), confirming that KIND2-F0Y13A lost the ability to interact with TLN1-F3.
To validate whether disrupting the talin–kindlin interaction influences binding to 15N-labeled β1-CT, we analyzed amide signal line widths in 1H–15N correlation spectra. The signals assigned to the α-helical region of β1-CT (from H758 to K770) showed little line broadening (intensity reduction of less than 10%) upon addition of TLN1-F3, ΔKIND2 or ΔKIND2Y13A at a molar stoichiometry of 1:0.8 compared to 1:0.2 (Fig. 2j), confirming that neither protein interacts with the α-helical region of the β1-CT (Fig. 1c and Extended Data Fig. 2n). By sharp contrast, signal intensities in the membrane-proximal α-helix decreased by 50% upon addition of TLN1-F3 together with ΔKIND2, by about 30% upon addition of TLN1-F3 together with ΔKIND2Y13A and by about 10% upon addition of TLN1-F3K402E together with ΔKIND2Y13A (Fig. 2j), indicating that disrupting the talin–kindlin interaction decreases but does not inhibit ternary interactions with β1-CT.
Talin and kindlin compete for β-CT without direct binding
The ability of KIND2Y13A to decrease ternary TLN1–β-CT–KIND2 complex formation assigns an important function to the talin–kindlin interaction that was tested with recombinant proteins in different experiments. First, we excluded structural changes in recombinant talin and kindlin that harbor the K402E or Y13A substitutions (Extended Data Fig. 3a–j). Subsequently, we measured their dissociation constants using MST, which revealed that THD1K402E was less potent (Kd,app,KIND2* = 16 ± 1 µM; Fig. 3a and Extended Data Fig. 3k) than THD1 in decreasing KIND2 affinity for β3-CT (Kd,app,KIND2* = 54 ± 27 µM; Fig. 1f,g) and that THD1 was less potent in decreasing KIND2Y13A affinity than KIND2 affinity for β3-CT (Kd,app,KIND2-Y13A* = 21 ± 3 µM; Fig. 3b and Extended Data Fig. 3l), indicating that the talin–kindlin interaction potentiates the decrease in kindlin affinity for the β3-CT. We also found that the KIND2Y13A-substituted protein gained the ability to decrease the affinity of THD1 for β3-CT (Kd,THD1 = 105 ± 20 µM; Kd,app,THD1* = 218 ± 55 µM; Fig. 3b and Extended Data Fig. 3n), indicating that the talin–kindlin interaction increases TLN1 affinity for the β3-CT, which, probably due to the low population of ternary talin–β-CT–kindlin complexes, escapes detection in MST measurements with wild-type THD1 and KIND2. Interestingly, insertions of the dissociation constants measured for THD1K402E, KIND2 and the β3-CT in the energy square produced a value of approximately 1, which agrees with an allosteric ternary-complex model (Fig. 3a):
as does THD1, KIND2Y13A and the β3-CT (Fig. 3b):
a,b, Schemes of MST-derived affinities of ternary-complex formation of THD1K402E and KIND2 (a) and THD1 and KIND2Y13A (b) with the β3-CT at thermodynamic equilibrium. c,d, FC-RDA-derived IC50 values of unlabeled THD1K402E and KIND2 competing for binding of 565–THD1K402E (blue and purple) and 647–KIND2 (orange and green) (c) or unlabeled THD1 and KIND2Y13A competing for binding of wild-type 565–THD1 (blue and purple) or 647–KIND2Y13A (orange and green) (d) to β1-TM–CT, β3-TM–CT and α5-TM embedded in PIP2- or PIP3-containing nanodiscs. e–h, MST affinity measurements of the TLN1-F3–ΔKIND2 fusion protein for 488–β1-CTY783A (e), β1-CTY795A (f) and β1-CT (g) and of the TLN1-F3–ΔKIND2Y13A fusion protein for 488–β1-CT (h). Data in e,f were fitted globally using a one-site-binding model resulting in Kd = 4.8 ± 1.1 µM for the talin-binding-deficient β1-CTY783A (kindlin contribution, green line) and Kd = 470 ± 140 µM for the kindlin-binding-deficient β1-CTY795A (talin contribution, blue line) and are shown for comparison as dashed lines in g,h. Data in g,h were fitted with two one-site-binding fits, setting the maximum response to the response fitted in e. Concentrations up to 5 µM were fitted with Kd = 2.9 ± 0.4 µM (g,h) (kindlin contribution, solid green line), whereas concentrations above 5 µM were fitted with Kd = 25 ± 3 µM (g) and Kd = 16 ± 1 µM (h) (talin contribution, solid blue line). i,j, MST affinity measurements of THD1 for 488–β1-CT (i) and 488–β3-CT (j) in the absence (blue) and the presence of 500–1,100 µM KIND2-F0 (yellow) or 300–400 µM β-CT-binding-impaired KIND2Q614A,W615A (QW/AA; orange). Data represent mean ± s.d.
The ability of KIND2 to compete with THD1K402E and the ability of KIND2Y13A to compete with THD1 for β-CT binding were confirmed by FC-RDA with β1-TM–CT and β3-TM–CT containing PIP2- or PIP3-supplemented nanodiscs (Fig. 3c,d). These data indicate that disruption of the talin–kindlin interaction leads to bidirectional competition of THD1 and KIND2 for β-CTs.
The low population of the ternary talin–β-CT–kindlin complexes, which further decreases in the absence of direct talin–kindlin interaction, impedes functional analysis of the ternary complex. To increase talin–kindlin binding events and thereby talin–β-CT–kindlin complex formation, we fused the C terminus of TLN1-F3 via a flexible GS linker to the N terminus of ΔKIND2 to produce the TLN1-F3–ΔKIND2 fusion protein and determined affinities for wild-type and mutant 488–β1-CTs. The affinities of TLN1-F3–ΔKIND2 for talin-binding-impaired β1-CTY783A and kindlin-binding-impaired β1-CTY795A were in good agreement with titrations of only KIND2 or only THD1 with β1-CT (Fig. 3e,f, Extended Data Fig. 1f,g and Extended Data Table 1). Furthermore, measurements revealed a positive, upward shift upon TLN1-F3–ΔKIND2 binding to β1-CTY783A (positive ΔFnorm values; Fig. 3e) and a negative, downward shift upon TLN1-F3–ΔKIND2 binding to β1-CTY795A (negative ΔFnorm values; Fig. 3f) in normalized MST traces, which we also observed in titrations of KIND2 and THD1 with β1-CT (Extended Data Fig. 1e,g). The opposing MST trends allowed us to distinguish the contributions of the talin–integrin and the kindlin–integrin binding site in titration experiments with the TLN1-F3–ΔKIND2 fusion protein with wild-type (WT) β1-CT (Fig. 3g). The experiment revealed that the titration of TLN1-F3–ΔKIND2 with β1-CT or β1-CTY783A produced the same kindlin-mediated upward shift in MST data and similar affinities (Kd,β1-CT-WT = 2.9 ± 0.4 µM; Kd,β1-CT-Y783A = 4.8 ± 1.1 µM; Fig. 3g). Of note, TLN1-F3-mediated ΔKIND2 competition remains invisible in titration experiments due to equimolar talin and kindlin concentrations in the TLN1-F3–ΔKIND2 fusion protein and the lower affinity of talin for β1-CT than the affinity of kindlin for β1-CT. In sharp contrast to the unaffected kindlin-mediated upward shift, the talin-mediated downward shift became strikingly steeper in the titration experiments, producing an 18-fold increase in affinity for β1-CT compared to β1-CTY795A (Kd,β1-CT-WT = 25 ± 3 µM; Kd,β1-CT-Y795A = 470 ± 140 µM; Fig. 3g). These data strongly indicate that kindlin is capable of increasing talin affinity for β1-CT when they both bind to the β1-CT.
To determine whether the association with kindlin induces allosteric activation of talin, followed by increased talin affinity for the β1-CT during ternary-complex formation, we titrated the TLN1-F3–ΔKIND2Y13A fusion protein with β1-CT (Fig. 3h). We observed identical binding curves as in titration experiments with the wild-type TLN1-F3–ΔKIND2 fusion protein binding to β1-CT (Fig. 3g,h). This result was confirmed in MST affinity measurements, which showed that THD1 affinity for β-CTs remained unchanged in the presence of β-CT-binding-deficient KIND2Q614A,W615A or KIND2-F0 (Fig. 3i,j), indicating that the association of TLN1 and KIND2 increases the population of the ternary talin–β-CT–kindlin complex but not THD1 affinity for β-CTs in the ternary complex.
Ternary-complex formation involves allostery in the β-CT
Because direct talin–kindlin interaction is not involved in the kindlin-mediated increase in talin affinity, we tested whether kindlin binding to the β-CT induces conformational changes that, in turn, increase β-CT affinity for talin. To test this hypothesis, we produced 2H,13C,15N-labeled β1-CT and partially deuterated TLN1-F3 and ΔKIND2 to record three-dimensional HNCACB spectra of β1-CT in the presence and the absence of 1:0.8 TLN1-F3 and/or ΔKIND2. From these spectra, we derived secondary 13Cα and 13Cβ chemical shifts (Δδ) that are indicative of secondary structure. The calculated Δδ13Cα–Δδ13Cβ values (Fig. 4a) are around 0 for disordered regions and are positive for α-helices and negative for β-sheets, with a maximum value of ~6 ppm indicating 100% secondary structure population23. In line with published structures4,11,24, 13C secondary chemical shifts of β1-CT assigned to membrane-proximal α-helical region were positive and regions around the KIND2-binding site indicate β-strand conformation, reflected by negative values. However, values of around |1| indicate only partial folding of the β1-CT in solution (Fig. 4a). Whereas addition of TLN1-F3 or ΔKIND2 alone induced minor changes, addition of both TLN1-F3 and ΔKIND2 induced 13C secondary chemical shifts pointing to increased α-helical conformation in the region between H758 and K774 and more extended β-type conformation in the region between W775 and V791. Compared to its unbound conformation, β1-CT is more structured in the presence of TLN1-F3 and ΔKIND2, suggesting that the ternary complex with talin and kindlin changes or selects a specific β1-CT conformation to enhance talin binding.
a, Secondary chemical shifts (Δδ13Cα–Δδ13Cβ) of 2H,13C,15N-labeled β1-CT in the absence (black, patterned) and the presence of partially deuterated ΔKIND2 (green), TLN1-F3 (blue) or ΔKIND2 and TLN1-F3 together (purple). Amino acid numbering is shown below plots. b, MST-derived binding affinities of THD1 and/or KIND2 for the β1-CT at thermodynamic equilibrium. The measured affinities are inconsistent with a simple ternary-complex model. c, Scheme of the β1–spacer–CT mutant. d, MST-derived binding affinities of THD1 and/or KIND2 for β1–spacer–CT at thermodynamic equilibrium. The observed affinities are consistent with a simple ternary-complex model.
To test the importance of allosteric coupling of the talin- and kindlin-binding sites (Fig. 4b), we separated the two binding sites by duplicating the intervening sequence with a ten-amino acid spacer (termed β1–spacer–CT; Fig. 4c) and found that neither THD1 changed KIND2 affinity nor KIND2 changed THD1 affinity for β1–spacer–CT (Fig. 4d and Extended Data Fig. 4a,b) and that the calculated dissociation constants complied with the ternary-complex model:
The independent binding of talin and kindlin to β1–spacer–CT increases ternary-complex formation, however, without inducing the structural conformational change in the integrin CT that is required for the function of the ternary talin–β-CT–kindlin complex.
Talin–kindlin cooperation is essential for cell adhesion
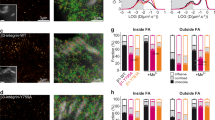
To test whether talin–kindlin interaction-mediated regulation of the population of ternary talin–β-integrin–kindlin complexes affects integrin function in cells, we retrovirally transduced mouse kidney fibroblasts lacking expression of TLN1, TLN2, KIND1 and KIND2 (ref. 1) (quadruple knockout (qKO)) to express N-terminally mCherry-tagged wild-type KIND2 or KIND2Y13A and C-terminally YPet-tagged wild-type TLN1 or TLN1K402E and subsequently sorted cell populations with similar TLN1–YPet and mCherry–KIND2 protein levels (qKO-TLN1WTKIND2WT, qKO-TLN1K402EKIND2WT, qKO-TLN1WTKIND2Y13A and qKO-TLN1K402EKIND2Y13A; Extended Data Fig. 5a–d) using flow cytometry. Whereas the total β1-integrin surface levels were alike on qKO-TLN1WTKIND2WT and qKO-TLN1K402EKIND2WT cells (Extended Data Fig. 5d), β1-integrin-activation-associated epitope 9EG7 levels were reduced on qKO-TLN1K402EKIND2WT cells before and after Mn2+ treatment (Fig. 5a). In line with the reduced β1-integrin activity, qKO-TLN1K402EKIND2WT cells produced fewer but on average larger adhesion sites, quantified by the mCherry–KIND2 signal (Fig. 5b,c), and showed slower adhesion strengthening, quantified by atomic force microscopy-based single-cell force spectroscopy (SCFS)25,26 (Fig. 5d,e), and impaired spreading on fibronectin (FN), laminin-111 and vitronectin (VN; Fig. 5f). Whereas qKO-TLN1WTKIND2WT cells responded with increasing cell size to increasing rigidity of FN-coated hydrogels, qKO-TLN1K402EKIND2WT cells retained their size on FN-coated hydrogels of 4-kPa, 12-kPa and 50-kPa rigidity (Fig. 5g). Importantly, qKO-TLN1WTKIND2Y13A, qKO-TLN1K402EKIND2WT and qKO-TLN1K402EKIND2Y13A cells displayed a similar phenotype. These findings were confirmed with an independent cell line3 expressing KIND2 from the endogenous Fermt2 locus, lacking expression of TLN1 and TLN2, and the defect was rescued with YPet-tagged wild-type TLN1 or TLN1K402E (double knockout (dKO)-TLN1; Extended Data Fig. 5a,c–f).
a, Binding of the β1-integrin-activation-reporting 9EG7 antibody in the presence of magnesium (white bars) or manganese (patterned bars) normalized to total β1-integrin levels. Dots represent experiments on different days (qKO, n = 5; dKO, n = 3; one-sample or two-sample t-test). b, Total internal reflection fluorescence (TIRF) microscopy images used for focal adhesion analysis of the indicated, serum-starved cells spread for 40 min on FN. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar, 10 µm. c, Average focal adhesion count and area per cell (n = 4 independent experiments; >8 cells per condition; one-way ANOVA of repeated measurements with Tukey’s post hoc test) quantified from the mCherry–KIND2 signal. d,e, qKO-TLN1WTKIND2WT (black) and qKO-TLN1K402EKIND2WT (blue) cells (d) and dKO-TLN1WT (gray) and dKO-TLN1K402E (red) cells (e) attached to a concanavalin A-coated cantilever brought in contact with FNIII7–10 for the indicated contact times and separated with a loading rate of 5 µm s−1. Dots represent separation forces of single fibroblasts, and lines indicate the median value. P values were calculated using two-tailed Mann–Whitney test and represent comparisons between qKO-TLN1WTKIND2WT and qKO-TLN1K402EKIND2WT cells or dKO-TLN1WT and dKO-TLN1K402E cells at the indicated contact times. f,g, Quantification of cell area from the TLN1–YPet signal 16 h after seeding serum-starved cells on FN, VN or laminin-111 (f) (laminin-111 (LN111), n = 3 independent experiments, >50 cells per condition) or FN-coated hydrogels with 4-, 12- or 50-kPa rigidity (g) (n = 3 independent experiments, >50 cells per condition analyzed, one-way ANOVA of repeated measurements with Tukey’s post hoc test). For details on statistical methods and data representation, see Methods.
To test the function of an integrin tail with decoupled talin- and kindlin-binding sites, we transduced β1-integrin complementary DNA encoding wild-type β1 or β1–spacer into β1-knockout fibroblasts (Extended Data Fig. 5g). Whereas flow cytometry revealed similar surface levels of total β1-integrin, cells expressing the β1–spacer displayed reduced levels of the integrin-activation-reporting epitope 9EG7 on their surface compared to cells expressing wild-type β1 (Extended Data Fig. 5h). Furthermore, FN-seeded β1–spacer-expressing cells exhibited profound adhesion and spreading defects (Extended Data Fig. 5i,j), which altogether indicates that formation of functional talin–β1-CT–kindlin complexes depends on the close proximity of talin–kindlin.
Discussion
Our NMR data indicate that β1-CT can assemble a ternary talin–β1-CT–kindlin complex. The ternary complex, which was shown to form also with the β3-CT and the β2-CT14,27, emerges as a principal molecular setting that ensures cooperativity between talin and kindlin, required to induce and maintain the active conformation of integrins. We expected that the two adaptor proteins augment each other’s binding to the β-CT rather than compete or bind independently (noncompetitively) of each other10. However, affinity measurements at equilibrium in solution and in the presence of charged lipid membranes point to a unidirectional competition mechanism, in which talin outcompetes kindlin from β1-CT or β3-CT, while kindlin does not affect talin binding. This observation agrees with single-particle tracking microscopy of live cells assigning kindlin a shorter immobilization time than talin in focal adhesions of FN-seeded fibroblasts28 and predicts three important consequences. First, the unidirectional competition leading to a drastic decrease in kindlin affinity for the β-CT indicates that the ternary talin–β-CT–kindlin complex is transient and rare. Second, unidirectional competition results from ternary talin–β-CT–kindlin complex formation. Third, assembly of the ternary talin–β-CT–kindlin complex is inconsistent with microscopic reversibility, suggesting that complex conformational change(s) are involved in the asymmetric binding behavior of talin and kindlin.
In search of such conformational changes, we first looked for a direct interaction between talin and kindlin that may influence their affinities for β-CTs. Although a direct interaction between talin and kindlin has not been reported thus far14, we detected, by NMR and cross-linking mass spectrometry, an interaction between the C-terminal α-helix of the TLN1-F3 domain and the N-terminal F0 domain of KIND2. Mutational disruption of the interaction decreased the population of ternary talin–β-CT–kindlin complexes by shifting the unidirectional competition toward bidirectional competition between talin and kindlin, in which kindlin decreases the affinity of talin and talin decreases the affinity of kindlin for β-CTs, although the latter less potently when compared with the competition studies in which the talin–kindlin interaction is intact. These findings indicate that, upon assembly of the ternary talin–β-CT–kindlin complex, kindlin induces a major increase in affinity of talin for the β1-CT and talin profoundly decreases kindlin affinity for the β1-CT. The increase in talin affinity for the β1-CT induced by kindlin could indeed be confirmed with a talin–kindlin construct connected by a flexible peptide linker.
Because β-CTs deficient for talin or kindlin binding excluded a role of direct talin–kindlin binding for inducing conformational changes in talin or kindlin that account for their asymmetric affinity behavior for the β-CT, we searched for a conformational communication of the talin- and kindlin-binding regions in the β1-CT that changes a low talin-affinity β-CT (termed ‘inactive’ β-CT) to a high talin-affinity β-CT (termed ‘active’ β-CT) conformation (Fig. 6a). We found secondary structure changes in the β1-CT region by NMR and could abolish allosteric coupling by introducing a spacer between the talin- and kindlin-binding sites that also abrogates asymmetric binding and curbs adhesion and spreading in cells. Interestingly, the population of ternary complexes is small, which is in line with single-molecule force measurements of RGD-bound αVβ3- and α5β1-integrins in cells29 that identified less than 10% of integrins in focal adhesions as transmitting high forces (exceeding 11 pN) and the remaining integrins, probably occupied only by either talin or kindlin, as transmitting low forces (below 11 pN).
a, Ternary complexes forming between talin, β-CT and kindlin in the presence or the absence of talin–kindlin binding. Because talin and kindlin compete for the inactive β-CT conformation, direct talin–kindlin interaction may help to overcome low talin affinity for the kindlin-occupied β-CT during ternary talin–β-CT–kindlin complex assembly. b, Talin–kindlin cooperativity involves a switch from an inactive to an active β-CT conformation. Kindlin interacts with inactive (bent-closed) and active (extended-open) integrins. Binding of kindlin to the extended-open integrin activates the β-CT, leading to high talin affinity and high resistance to actomyosin pulling forces. High talin affinity in turn decreases kindlin affinity for the β-CT, resulting in kindlin dissociation from the β-CT, β-CT inactivation, a decrease in talin affinity and the integrin bent-closed state. Talin–kindlin interaction at the β-CT maintains a low population of ternary talin–β-CT–kindlin complexes by helping to overcome low talin affinity for the kindlin-occupied β-CT until the active β-CT conformation is induced and the ternary complex is assembled. The low population of ternary complexes in focal adhesions is able to transmit high forces, while the remaining integrins, occupied by only talin or kindlin, transmit low forces.
THD1 is a FERM domain, and its crystal structure revealed both an atypical, linear and a canonical, cloverleaf-like conformation5,30. The cloverleaf-like conformation displays an interdomain interaction between D125 and E126 of TLN1-F1 and K401 and K402 of the TLN1-F3 domain, the latter of which is directly adjacent to the talin–kindlin interaction site identified in our study (Extended Data Fig. 6). It is conceivable that the linear and cloverleaf conformations of THD1 are in equilibrium, and a preference of KIND2 binding for the linear TLN1 conformation might increase membrane affinity via accessibility of additional membrane contacts by the TLN1-F0 and TLN1-F1 domains. This hypothesis is supported by our FC-RDA experiments, which showed that unidirectional talin-mediated kindlin competition is more pronounced from β1-TM–CT- and β3-TM–CT-containing nanodiscs than from β-CTs in solution. Hence, the interaction between talin and kindlin may also promote talin activation by increasing talin affinity for the plasma membrane, whereas interactions occurring simultaneously between talin, β-CT and kindlin increase talin affinity for the β-CT.
Our data identify a complex mechanism underlying talin and kindlin cooperativity (Fig. 6b). Based on experimental evidence, we envision that talin–kindlin cooperativity commences with kindlin, which encounters the integrin before talin31. Because talin and kindlin compete for the inactive β-CT conformation, direct talin–kindlin interaction may help to overcome low talin affinity for the kindlin-occupied β-CT and keep talin close to the β-CT until the active β-CT conformation is induced. Binding of talin and kindlin induces the ‘active’ β-CT conformation, which increases talin affinity for the β-CT, reinforces talin–β-CT binding and enhances resistance to actomyosin pulling forces. The fascinating feature of the asymmetric binding behavior is that it is inherently autonomous: elevated talin affinity for β-CT results in a decrease in kindlin affinity for the active β-CT conformation, leading to kindlin unbinding, a decrease in talin affinity and eventually talin dissociation, integrin inactivation and initiation of a new cycle of ternary talin–β-CT–kindlin assembly.
Methods
Cloning
Deletions, mutations and short insertions in complementary DNA were introduced by PCR using PfuUltra II (600670, Agilent Technologies) or Q5 Hot Start polymerase (M0493, New England Biolabs) according to the manufacturer’s protocol, followed by DpnI (R0176, New England Biolabs) digestion of template DNA (2 h, 37 °C). After cleanup (QIAquick PCR Purification Kit, 28104, Qiagen), the PCR product was phosphorylated using T4 PNK (M0201, New England Biolabs), followed by another cleanup, ligated (Fast-Link DNA Ligation Kit, Epicenter) for 30 min at room temperature and transformed into Escherichia coli OmniMAX (Promega) or NEB 5-alpha competent E. coli (New England Biolabs).
Long insertions were introduced using the NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs, E5520S) according to the manufacturer’s protocol. The assembled constructs were transformed into E. coli OmniMAX or NEB 5-alpha competent E. coli.
DNA encoding the β1A-CT was synthesized by Eurofins Genomics.
Protein expression and purification
Kindlin-2
KIND2 and ΔKIND2 were expressed and purified as described earlier1. Briefly, kindlin constructs were cloned into pCoofy17 (ref. 32), which adds an N-terminal His10–SUMO tag and expressed in soluble form in E. coli Rosetta cells at 18 °C overnight. After purification by immobilized metal chelate affinity chromatography (IMAC) in high-salt TBS buffer (20 mM Tris, pH 7.5, 500 mM NaCl, 1 mM Tris(2-carboxyethyl)phosphine (TCEP)), the SUMO tag was removed by SenP2 protease (obtained from the Max Planck Institute of biochemistry (MPIB) Core Facility) digest overnight, and the protein was further purified by SEC using TBS (20 mM Tris, pH 7.5, 200 mM NaCl, 1 mM TCEP) containing 5% glycerol as the running and storage buffer.
Talin
DNA for the THD and F3 domains of TLN1 (1–405 and 305–405) and TLN2 (1–408 and 311–408) was cloned into pCoofy17, which adds an N-terminal His10–SUMO tag. After induction with isopropyl β-d-1-thiogalactopyranoside (IPTG) at an OD600 of 0.6, proteins were expressed in E. coli (DE3) Rosetta 2 cells grown in TB, LB or M9 medium at 18 °C for 18–24 h. Cells were collected by centrifugation (6,000g, 20 min, 4 °C), resuspended in lysis buffer (20 mM Tris, pH 7.5, 500 mM NaCl, 1 mM TCEP, lysozyme, benzonase), disrupted by sonication (1-s pulses, 10 min, 90% amplitude) or high-pressure homogenization (Avestin Emulsiflex C3, ATA Scientific) and centrifuged (58,000g, 20 min, 4 °C), and the supernatant containing soluble proteins was filter sterilized (0.22 µm) and subjected to IMAC (HisTrap FF, GE Healthcare), followed by elution with a stepwise gradient. Fractions containing the protein (typical elution with 250–500 mM imidazole) were pooled, concentrated and rebuffered against TBS (20 mM Tris, pH 7.5, 200 mM NaCl) to remove imidazole before adding 40–80 µl SenP2 protease and incubating at 4 °C overnight. The cleaved His–SUMO tag, uncleaved protein and the SenP2 protease were removed by incubating the sample at 4 °C for 30 min while rotating with Ni-NTA beads. The sample was filtered (0.22 µm) and purified by SEC using a Superdex 75 10/300 GL, a Superdex 200 10/300 GL or a HiLoad Superdex 200 PG column (GE Healthcare), depending on molecular weight and purification scale.
α-TM–CTs and β-TM–CTs
Integrin TM–CTs were expressed and purified as described earlier with slight variations33. The β1-TM–CT (726–798), the β3-TM–CT (715–787), the mutant β1-TM–CT (W775A/Y783A, T778A/T789A/Y795A) and α5-TM (993–1,025) constructs were produced in E. coli BL21 (DE3) Rosetta 2 cells, leading to the formation of inclusion bodies. The cells were resuspended in lysis buffer (50 mM HEPES, 150 mM NaCl, 0.1% Triton X-100, pH 7.5, 1 mM TCEP, 1 mM PMSF, lysozyme, benzonase), disrupted by sonication (1-s pulses, 10 min, 90% amplitude) and centrifuged (58,000g, 20 min, 4 °C), and supernatant containing soluble proteins was discarded. The sediment was washed once with 50 mM HEPES, 150 mM NaCl, 0.1% Triton X-100, pH 7.5, followed by another centrifugation (58,000g, 20 min, 4 °C), and resuspended in 50 mM HEPES, 150 mM NaCl, 3% EMPIGEN (β-TM–CT) or 3% n-dodecyl-β-d-maltopyranoside (α5-TM), 1 mM TCEP, pH 7.5 to solubilize the TM–CT constructs from inclusion bodies. The sample was incubated overnight on a rotating wheel at 4 °C, except for mutant β1-TM–CTs, which were incubated for 2 d at 37 °C. After centrifugation (58,000g, 20 min, 4 °C), the supernatant was filter sterilized and purified by IMAC (HisTrap FF, GE Healthcare) and cation-exchange chromatography (HiTrap SP HP, GE Healthcare) at pH 5.8.
Integrin-β cytoplasmic tails
The integrin-β1-CT (758–798) was cloned into pCoofy17, which adds an N-terminal His10-SUMO tag. After induction with IPTG at an OD600 of 0.6, proteins were expressed in E. coli (DE3) Rosetta 2 cells in M9 medium at 18 °C for 36 h. Cells were collected by centrifugation (6,000g, 20 min, 4 °C), resuspended in lysis buffer (20 mM Tris, pH 7.5, 500 mM NaCl, 1 mM TCEP, lysozyme, benzonase), disrupted by sonication (1-s pulses, 10 min, 90% amplitude) and centrifuged (58,000g, 20 min, 4 °C); the supernatant containing soluble proteins was filter sterilized (0.22 µm) and subjected to IMAC (HisTrap FF, GE Healthcare), followed by elution with a stepwise gradient. Fractions containing the protein (typical elution with 250–500 mM imidazole) were pooled, diluted with high-salt TBS buffer and concentrated by another IMAC (HisTrap FF, GE Healthcare) with a single elution step using high-salt TBS buffer with 500 mM imidazole. The resulting elution fraction was diluted tenfold with 20 mM Tris-HCl, pH 7.0, before adding 80 µl SenP2 protease and incubating at 4 °C overnight. The cleaved His–SUMO tag, uncleaved protein and the SenP2 protease were removed by incubating the sample at 4 °C for 30 min while rotating with Ni-NTA beads. The sample was filtered (0.22 µm), and the pH value was adjusted to about 6 using 1 M MES, pH 5.8 and confirmed with pH paper. Next, the protein was loaded on a cation-exchange chromatography column (HiTrap SP, 5 ml, GE Healthcare), washed with 25 mM MES, pH 5.8, 40 mM sodium chloride buffer and eluted with TBS (20 mM Tris, pH 7.5, 200 mM sodium chloride, 1 mM TCEP). Purity and labeling efficiency with 13C and 15N isotopes were determined by mass spectrometry and were >98%.
After the final chromatography step, the purity, integrity and identity of recombinant KIND2 and talin proteins were controlled by SDS–PAGE, high-resolution mass spectrometry and dynamic light scattering (DLS). Integrin TM–CT peptides were controlled by SDS–PAGE and high-resolution mass spectrometry. Peptides were controlled by high-resolution mass spectrometry.
Fluorescent labeling and biotinylation of proteins
Before labeling, proteins were transferred into a buffer suitable for the intended labeling reaction using desalting columns. Integrin CTs were labeled with ATTO 488 N-hydroxysuccinimide (NHS) at their N terminus during chemical synthesis (by the MPIB core service facility), integrin TM–CTs were labeled with biotin-maleimide, whereas kindlin and talin were labeled with ATTO 565 NHS or Alexa 647 NHS or maleimide. For maleimide labeling, the pH value was adjusted to 7.0–7.5 using 1 M Tris, pH 7.5, and cysteines were reduced by adding TCEP to a final concentration of 2 mM before the reaction. Next, thiol-reactive dye was added at a molar excess of 10–20× and incubated at room temperature for 2 h in the dark or at 4 °C overnight. For amino-reactive dyes, the protein was first transferred to NHS labeling buffer (PBS with 10 mM NaHCO3, pH 9.0) and then mixed with dye at a molar excess of 2.5×. The labeling reaction was carried out for 1 h at room temperature or overnight on ice in the dark. Excessive dye was removed with desalting columns.
Assembly of nanodiscs
Lipids were dissolved in chloroform (1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC)), 20:9:1 chloroform–methanol–water (PIP2) or 80:40:1 chloroform–methanol–HCl (PIP3) at a final concentration of 50 mg ml−1 (DMPC) or 10 mg ml−1 PIP2 or PIP3, which yielded a final concentration of 10% PIP2 and 90% DMPC or 10% PIP3 and 90% DMPC to obtain molar ratios of 1:9 PIP2–DMPC and 1:9 PIP3–DMPC, respectively. The desired volume of lipid solution was transferred to a clean glass tube using a Hamilton glass syringe, dried under an N2 stream, followed by desiccation overnight, and dissolved in cholate buffer (20 mM Tris, pH 7.5, 100 mM sodium chloride, 100 mM sodium cholate) to yield a 50 mM lipid stock solution.
For nanodisc assembly, integrin TM–CT, MSP2N2 scaffold protein (obtained from the MPIB Biochemistry Core Facility) and lipid stock solution were mixed 1:1:330 to obtain on average one integrin TM–CT per nanodisc. Before adding lipids to the mixture, cholate buffer was added to obtain a final cholate concentration between 10 and 20 mM and to avoid precipitation of lipids. The samples were then dialyzed at least three times against 1 l of nanodisc buffer (20 mM HEPES, pH 7.5, 150 mM sodium chloride, 0.5 mM EDTA) at room temperature, filter sterilized and purified with a Superdex 200 Increase 10/300 GL (GE Healthcare) or an SEC 650 (Bio-Rad) column to separate assembled nanodiscs from non-assembled components. The elution fractions were analyzed by SDS–PAGE with silver staining and flow cytometry for talin and/or kindlin binding.
Microscale thermophoresis measurements
All MST measurements were performed as published34 on a Monolith NT.115 red–blue machine (NanoTemper) using premium coated capillaries to reduce nonspecific interaction of proteins with the glass surface. Both interaction partners (ligand and receptor) were transferred into MST buffer (20 mM Tris, pH 7.5, 200 mM sodium chloride, 1 mM TCEP, 0.05% Tween-20) to avoid artifacts derived from buffer mismatches. ATTO 488-labeled integrin-β-CTs (50–200 nM, synthesized by the MPIB Core Facility) were used as ligands. Measurements were carried out at 10–20% LED power and 20% and 40% MST power. Data were analyzed using MO.Affinity Analysis Software (NanoTemper) as shown in Extended Data Fig. 1e. MST figures display data from individual titrations that were pooled (circles) and fitted with a global one-site-binding curve (lines). Affinity data in the figure insets (bar charts), in Extended Data Table 1 and in the text are mean ± s.d. of the replicates, which are given as n.
Flow cytometry-based reporter-displacement assay
Fluorescently labeled 565–THD1 or 647–KIND2 were dissolved at a final concentration of 100 nM and 50 nM, respectively, in 200 µl 20 mM Tris, pH 7.5, 200 mM sodium chloride containing 0.25 µl Dynabeads M-280 Streptavidin (11205D, Invitrogen). To this buffer, competitors, that is, unlabeled THD1 and KIND2, were titrated before adding 10 µl of biotinylated integrin α-TM–CT or β-TM–CT in nanodiscs and incubating the solution for 10 min at room temperature while rotating. The samples were then injected in an LSRFortessa X-20 flow cytometer (BD Biosciences), and data were analyzed with FlowJo 10 (FlowJo) and OriginPro 2019b (OriginLab).
For data analysis, beads were gated in FlowJo and used as the negative control without protein and nanodiscs. MFIs of the samples were determined and exported to OriginPro 2019b. Here, data were fitted to a dose–response curve, setting top and bottom asymptotes to the values measured for the positive and negative controls, respectively. The positive control was measured in the presence of nanodiscs but in the absence of competitor, while, in the negative control, competitor and nanodiscs were absent. We report IC50 values of these fits of individual experiments with different nanodisc preparations, which were recorded on different days as mean ± s.d.
Dynamic light scattering
Before conducting a DLS measurement, protein samples were centrifuged for 15 min at 21,000g and 4 °C. DLS measurements were performed in triplicate on a DynaPro NanoStar instrument (Wyatt) at 20 °C with laser power set to auto-attenuation, an acquisition time of 5 s and 15 acquisitions. The hydrodynamic radius of the particles in the sample was calculated with Dynamics software (Wyatt).
Circular dichroism spectroscopy
CD spectra were acquired using a quartz cuvette with a path length of 1 cm and a sample volume of 300 µl in a Jasco J-715 spectropolarimeter. Before the measurements, a buffer reference and protein samples (concentration, 0.1 mg ml−1) were prepared with PBS buffer. First, a CD spectrum of the buffer sample was acquired from 190 nm to 250 nm at 25 °C. Afterward, buffer was removed from the cuvette, and the protein sample was added and measured using the same parameters. The buffer reference spectrum was subtracted from the protein spectrum to eliminate effects caused by the buffer.
Thermal stability
Thermal stability was measured on a Prometheus NT.48 (NanoTemper) using a temperature gradient from 20 to 95 °C with an increase in temperature of 1 °C min−1 while measuring internal tryptophan fluorescence at λ = 330 nm.
NMR spectroscopy
For triple-resonance experiments with the β1-CT, M9 medium was supplemented with 0.5 mg ml−1 [15N]ammonium chloride or with 2 mg ml−1 [13C]glucose. All experiments were performed in NMR buffer consisting of 20 mM Tris, 200 mM NaCl, 1 mM DTT, pH 7.5 and 5–10% D2O for the NMR lock signal. For deuterated β1-CT samples, cells were adapted to deuterated M9 minimal medium in steps of 0%, 50% and 80% D2O supplemented with 0.5 mg ml−1 [15N]ammonium chloride and 2 mg ml−1 [13C]glucose, whereas ΔK2- and T1-F3-expressing cells were grown in 67% D2O M9 minimal medium. All datasets were acquired from Bruker Avance III spectrometers at a proton frequency of 600–800 MHz equipped with triple-resonance cryoprobes using TopSpin 3.2–3.5 software. Data were processed with TopSpin or NMRPipe and analyzed using CcpNmr Analysis software. Sample concentrations ranged from 70 µM to 700 µM, with all NMR experiments carried out at 298 K.
Protein backbone resonance assignments of β1-CT were obtained using 3D HNCO, HNcaCO, HNCACB and CBCAcoNH experiments. Assignments for β1-CT methyl resonances were performed using 3D (H)C(CCO)NH and H(CCCO)NH. Titrations were performed by forming binary or ternary complexes of either 15N- or 13C,15N-isotope-labeled integrin β1-CT mixed with unlabeled TLN1-F3 and ΔKIND2 at the indicated stoichiometries. For each titration point, 1H–13C constant time HSQC or 1H–15N HSQC spectra were recorded.
For NMR titration measurements, wild-type TLN1-F3 and the TLN1-F3K402E mutant were expressed in M9 medium supplemented with 0.5 mg ml−1[15N]ammonium chloride. Backbone assignments for wild-type TLN1-F3 were transferred from a previously published study in the Biological Magnetic Resonance Data Bank (BMRB 7061). Titrations were performed by forming binary or ternary complexes of 15N-labeled wild-type TLN1-F3 or TLN1-F3K402E protein with unlabeled β1-CT or ΔKIND2 at the indicated stoichiometry. For KIND2-F0, assignments were transferred from the Biological Magnetic Resonance Data Bank (BMRB 30659). About 47% of the backbone amide chemical shifts could be transferred by comparing 1H–15N correlation spectra. Other signals exhibited some chemical shift differences or could not be unambiguously identified (Supplementary Table 1). Titrations involving wild-type KIND2-F0 or KIND2-F0Y13 were performed at the indicated stoichiometries. For all measurements, 1H–15N HSQC spectra were recorded at each point of the titration, the chemical shift changes of amide resonances in the fast-exchange regime were measured, and the reported weighted-average values of 15N and 1H chemical shift changes are given by equation (3):
Cross-linking of proteins
The TLN1-F3–β1-CT fusion protein was mixed with KIND2 at a molar excess of 7.5-fold and concentrated to a final concentration of 60 µM TLN1-F3–β1-CT and 450 µM KIND2 in 120 µl TBS. The protein mixture was rebuffered against PBS using a 0.5-ml Zeba Spin column according to the manufacturer’s protocol and subjected to chemical cross-linking using the GraFix method as described earlier35. In brief, 5–20% sucrose gradients were generated in SW 40 ultracentrifuge tubes using a Gradient Master station (model IP, Biocomp) with 0.5 mM DSS in the heavy solution. Concentrated proteins were added to the tubes on top of the gradients, and the setup was centrifuged in an SW 40 Ti swing bucket rotor at 40,000 r.p.m. for 16 h (Beckman Coulter). After centrifugation, the gradients were fractionated on the Gradient Master station coupled to a Bio-Rad fraction collector, and fractions were analyzed for complex-containing fractions by SDS–PAGE and Coomassie staining. Fractions of interest were analyzed by mass spectroscopy.
Cross-linking mass spectrometry
Cross-linked protein pellets were incubated in digestion buffer (1:1; 1% SDC, 40 mM CAA, 10 mM TCEP, 50 mM Tris) for 20 min at 37 °C and then diluted with water (VWR) and finally digested at 37 °C overnight with 2 µg trypsin (Promega). The peptide mixture was acidified, desalted with Sep-Pak C18 1 cc vacuum cartridges (Waters), dried in a vacuum and dissolved in buffer A (0.1% formic acid, at a concentration of 400 ng µl−1). The peptides (400 ng) were separated with the Thermo EASY-nLC 1200 System (Thermo Fisher Scientific; flow rate of 250 nl min−1), equipped with a 30-cm analytical column (inner diameter, 75 µm; packed in house with ReproSil-Pur C18-AQ 1.9-µm beads, Dr. Maisch) coupled to the benchtop Orbitrap Q Exactive HF (Thermo Fisher Scientific) mass spectrometer, with an increasing gradient of buffer B (80% acetonitrile, 0.1% formic acid).
The raw data were processed with Proteome Discoverer (version 2.5.0.400) with XlinkX/PD nodes integrated36. DSS or BS3 was set as a cross-linker, cysteine carbamidomethylation was set as a fixed modification, and methionine oxidation and protein N-terminal acetylation were set as dynamic modifications. ‘Trypsin/P’ was specified as the protease, and up to two missed cleavages were allowed. Identifications were only accepted with a minimal score of 40 and a minimal delta score of 4. Filtering at a false discovery rate of 1% was calculated with the XlinkX Validator node with the setting ‘simple’.
Generation of the structural model
As structural data are unavailable for critical regions of the proteins used in our study, we generated the structural model manually. To this end, we used the crystal structure of the β1D-CT–TLN2 complex (PDB 3G9W) because the structures of neither β1A-CT nor β1A-CT in complex with TLN1 (β1A-CT–TLN1) have been solved yet. Because the kindlin-binding site of β1D-CT differs from that of β1A-CT and is not resolved in the structure, we used the β1A-CT–ΔKIND2 crystal structure (PDB 5XQ0) and aligned the resolved amino acids of β1A-CT–ΔKIND2 and β1D-CT–TLN2. Furthermore, structural information of the flexible N terminus of the KIND2-F0 domain is also not resolved in any of the published KIND2 crystal structures. Therefore, we aligned the NMR structures of the individual KIND2-F0 domain (PDB 6U4N) with the model. Subsequently, we mapped the intramolecular and intermolecular cross-links obtained in our cross-linking mass spectrometry experiments onto these assembled published structures (TLN2-F2F3–β1D11, ΔKIND2–β1A4 and KIND2-F0 (ref. 37)) using the XMAS plugin38 for ChimeraX39. We colored cross-links of the relevant distance of 5–30 Å in yellow and all remaining cross-links in red and then iteratively adjusted the orientation of TLN1, KIND2 and β1-CT toward each other until the maximal number of cross-links were of the relevant distance.
Cell culture
Cells were grown and maintained in DMEM medium supplemented with 10% FBS and 1% penicillin–streptomycin on 10-cm Petri dishes. At about 80% confluency, cells were washed with 5 ml PBS and detached with 1 ml 0.05% trypsin–EDTA in PBS at room temperature before adding 5 ml warm DMEM with 10% FBS. Cells were sedimented by centrifugation (5 min, 350g, room temperature), the supernatant was removed, and the cells were resuspended in 5 ml warm DMEM with 10% FBS. Cells were stained with Trypan blue to determine cell count and viability (EVE, NanoEnTek). Next, 200,000–400,000 cells were seeded in 10 ml DMEM with 10% FBS and 1% penicillin–streptomycin in 10-cm Petri dishes. All applied cell lines regularly tested negative for mycoplasma contamination.
The plasmid encoding murine TLN1–YPet in the retroviral vector pLPCXmod has been described previously40. The TLN1K402E mutation was introduced by site-specific mutagenesis as described in Cloning. The plasmid encoding mCherry–KIND2 is based on EGFP–KIND2 in the retroviral vector pRetroQ-AcGFP-C1 (Clontech) as published earlier1. The plasmid encoding human β1-integrin in the retroviral vector pLZRS was described earlier41.
Retroviruses for stable transduction were grown in HEK293T cells, and Tln1−/−;Tln2−/−-dKO, Tln1−/−;Tln2−/−;Fermt1−/−;Fermt2−/−-qKO and Itgb1−/− murine kidney fibroblasts were generated and transduced as published earlier1,3,41. After transduction, dKO cells rescued with wild-type TLN1–YPet and TLN1–YPetK402E were sorted for equal YPet MFI signal, qKO cells rescued with wild-type TLN1–YPet and TLN1–YPetK402E and wild-type mCherry–KIND2 and mCherry–KIND2Y13A were sorted for equal YPet and mCherry MFI signals, and rescued Itgb1-knockout cells were sorted for equal PE MFI signal after staining for total β1-integrin surface levels with a PE-labeled anti-β1-integrin antibody using the FACSAria III Cell Sorter (BD Biosciences). Talin and kindlin expression levels of wild-type TLN1–YPet and TLN1–YPetK402E dKO cells and wild-type TLN1–YPet and TLN1–YPetK402E qKO cells were further compared to those of Tln1fl/fl;Tln2−/−;Fermt1fl/fl;Fermt2fl/fl fibroblasts by western blot following our previous publication1.
Flow cytometry
Around 400,000 cells were seeded into a well of a six-well cell culture plate the day before performing flow cytometry. The cells were detached from the culture plates with 500 µl trypsin and EDTA in PBS, trypsin was neutralized with 500 µl DMEM medium supplemented with 10% FBS, and samples were transferred into 4 ml DMEM supplemented with 10% FBS and split into different FACS tubes or 96-well plates. The medium was removed by centrifugation (5 min, 350g, 4 °C), and cells were washed twice with cold PBS and incubated with primary antibodies (9EG7 monoclonal antibody, which binds extended β1-integrin (1:100), or total β1-integrin (1:200), diluted in adhesion buffer (PBS, 3% BSA, 4.5 g l−1 glucose, 1 mM calcium chloride, 1 mM magnesium chloride)) for 30 min on ice. After washing with cold PBS, secondary antibodies (anti-rat 647 and streptavidin-720) were diluted 1:500 in adhesion buffer and added to cells for 30 min on ice. The cells were washed again with cold PBS and suspended in PBS supplemented with 3% BSA. For integrin profiling, cells were incubated after washing with PBS at a 1:50 dilution of PE-labeled anti-β1-integrin, anti-β3-integrin, anti-α5-integrin, anti-αV-integrin antibodies or the corresponding isotype controls in PBS supplemented with 3% BSA for 15 min at room temperature. All measurements were performed with an LSRFortessa X-20 flow cytometer (BD Biosciences). Data were analyzed with FlowJo 10 and OriginPro 2019b. Binding of the 9EG7 antibody was normalized to total β1-integrin levels (9EG7 signal divided by total β1 signal). For integrin profiling, the MFI for each integrin staining was first corrected for its corresponding isotype control, and then data from TLN1–YPetK402E dKO and qKO cells were normalized to wild-type TLN1–YPet dKO and qKO cells. Each data point shown originates from an experiment on an individual day. Bar charts show mean ± s.d.
Focal adhesion analysis
Eight-well glass slides (tissue culture treated, 0030 742.036, Eppendorf) were coated with 5 µg ml−1 FN (341635, Merck) in PBS for at least 30 min at 37 °C, followed by blocking with 3% BSA in PBS for at least 30 min at 37 °C. After washing the wells with PBS and DMEM, 5,000–10,000 cells serum-starved for 4 h in DMEM were seeded and incubated for 40 min at 37 °C. The medium was removed, and the cells were fixed with 4% paraformaldehyde (PFA) in PBS for 10 min at room temperature, followed by DAPI staining (1:10,000 in PBS) for 5 min at room temperature and three washing steps with PBS. The cells were imaged on a custom-made TIRF microscope (VisiTIRF, Visitron Systems) based on an Observer Z1 microscopy stand (Carl Zeiss). TIRF illumination was performed with a ×100 TIRF objective (Plan-Apochromat ×100/1.46 Oil, Carl Zeiss). Focal adhesion properties were analyzed based on TIRF images recorded in the 561-nm laser channel for the mCherry–KIND2 signal using the Focal Adhesion Analysis Server, setting the threshold to 2 and the minimal adhesion size to five pixels42.
Spreading
Eight-well slides (µ-Slides, 80826, ibiTreat, ibidi) or six-well plates with hydrogels of different stiffnesses (Cell Guidance Systems) were coated with 5 µg ml−1 FN (341635, Merck) in PBS, 2.5 µg ml−1 VN (07180, Stemcell Technologies) or 10 µg ml−1 laminin-111 in PBS (L2020, Sigma-Aldrich) for at least 30 min at 37 °C, followed by blocking with 3% BSA in PBS for at least 30 min at 37 °C. After washing the wells with PBS and DMEM, 5,000–10,000 cells serum-starved for 4 h in DMEM were seeded and incubated for different times. When working with eight-well slides, the medium was afterward removed, and the cells were fixed with 4% PFA in PBS for 10 min at room temperature, followed by DAPI staining (1:10,000 in PBS) for 5 min at room temperature and three washing steps with PBS. When working with six-well plates and hydrogels, cells were incubated for 24 h, washed with PBS and immediately imaged to avoid artifacts from overfixation. Cells were imaged on an EVOS FL Auto 2 microscope (Invitrogen). The area of at least 50 individual cells was determined with Fiji ImageJ using the fluorescent signal of the reconstituted TLN1–YPet constructs.
Single-cell force spectroscopy
For cantilever functionalization, cantilevers (NP-0, Bruker) were first plasma cleaned (PDC-32G, Harrick Plasma) and then incubated overnight in PBS containing concanavalin A (2 mg ml−1, Sigma-Aldrich) at 4 °C43. For substrate coating, 200-µm-thick four-segmented polydimethylsilane masks were fused to glass surfaces of Petri dishes (WPI)44. Polydimethylsilane-framed glass surfaces were incubated overnight with an FN fragment (FNIII7–10RGD, 50 µg ml−1 in PBS) at 4 °C. A NanoWizard II AFM equipped with a CellHesion module (both from JPK Instruments) mounted on an inverted fluorescence microscope (Observer Z1, Zeiss) was used for SCFS. The temperature was controlled at 37 °C by a PetriDishHeater (JPK Instruments). Tipless V-shaped silicon nitride cantilevers (200 µm long, NP-0) having nominal spring constants of 0.06 N m−1 were used. Each cantilever was calibrated before measurement by determining its sensitivity and spring constant using the thermal noise analysis of the AFM.
Fibroblasts were grown to a confluency of ~80%, washed with PBS, detached with trypsin for 2 min, suspended in SCFS medium (bicarbonate-free DMEM supplemented with 20 mM HEPES) containing 1% (vol/vol) FCS and centrifuged, and the sedimented cells were resuspended in serum-free SCFS medium. Fibroblasts were allowed to recover from trypsin detachment for at least 30 min45. Afterward, suspended fibroblasts were pipetted onto substrate-coated Petri dishes, and the functionalized cantilever was lowered onto a single fibroblast with a speed of 10 µm s−1 until a force of 5 nN was recorded. After 5 s of contact, the cantilever was retracted at 10 µm s−1 until the cell was completely detached from the substrate, and the cantilever-bound fibroblast was incubated for 3–5 min on the cantilever to ensure firm binding to the cantilever. The morphological state of the fibroblast was monitored by optical microscopy throughout adhesion experiments. Round, cantilever-bound fibroblasts approached the substrate at 5 µm s−1 until a contact force of 1 nN was recorded. Throughout the contact times of 5, 20, 60, 120 or 240 s, the height of the cantilever was maintained constant and subsequently retracted at 5 µm s−1 for 100 µm until the fibroblast was fully separated from the substrate. Before another experimental cycle was initiated, the cantilever-bound fibroblast was allowed to recover for a time period equal to the contact time. A single fibroblast was used to probe the adhesion force for all contact times once or until morphological changes (that is, spreading) were observed. The sequence of contact times and area on the substrate were varied. Adhesion forces were determined from force–distance curves as the maximum downward deflection of the cantilever after baseline correction using JPK software (JPK Instruments).
Plate-and-wash assay
Untreated 96-well plates were coated with 100 µl of 10 µg ml−1 FN or a 1:10 dilution of poly-l-lysine (Sigma) in 20 mM Tris, pH 8.8, overnight at 4 °C. Next, 200 µl of 3% BSA in PBS was added to the wells for at least 30 min at 37 °C. The plates were washed with 200 µl PBS and 200 µl DMEM medium before seeding 40,000 serum-starved cells in 100 µl DMEM. After incubating for 30 min at 37 °C, the whole 96-well plate was washed by immersing in PBS three times, followed by fixation in 4% PFA for 10 min at room temperature. The cells were stained with 75 µl crystal violet solution for 30 min at room temperature and washed three times by immersing with dH2O. The wells were incubated with 100 µl 2% SDS while shaking at room temperature, until all crystal violet was dissolved. Absorption was measured at λ = 595 nm in a SpectraMax ABS plate reader (Molecular Devices). The data for every cell line were first blanked to those of control wells coated with only BSA and then corrected for the plated cell count obtained from poly-l-lysine-coated wells. Finally, data were normalized to those of the wild-type β1-expressing cell line.
Statistics and data representation
Two samples were compared to each other using a two-sample two-tailed Student’s t-test (Figs. 1e,f, 3l,j and 5a and Extended Data Figs. 1g,h,l,j,m, 3k–n and 4a,b) or a two-tailed Mann–Whitney test (Fig. 5d,e). For comparisons with more than two samples, one-way repeated-measures ANOVA with Tukey’s post hoc test was used (Fig. 5c,f,g and Extended Data Fig. 5e,f,I,j). Normalized data were tested with a one-sample two-tailed Student’s t-test for a difference from 1 for comparison with the reference sample (Fig. 5a and Extended Data Fig. 5d,h,i). P values from statistical tests are written in the figures over the corresponding bar chart or box plot in black, whereas n is written in the same color as the chart or plot. Unless stated otherwise, bar charts represent average values and error bars represent standard deviations of individual experiments or analyzed single cells. In box plots, the boxes show the upper and lower quartiles and the median and the error bars show standard deviations of individual experiments.
Reagents
Reagents used in the study are detailed in Table 1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Additional data and materials can be obtained from the corresponding author upon request. Source data are provided with this paper.
References
Bottcher, R. T. et al. Kindlin-2 recruits paxillin and Arp2/3 to promote membrane protrusions during initial cell spreading. J. Cell Biol. 216, 3785–3798 (2017).
Moser, M., Legate, K. R., Zent, R. & Fassler, R. The tail of integrins, talin, and kindlins. Science 324, 895–899 (2009).
Theodosiou, M. et al. Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. eLife 5, e10130 (2016).
Li, H. et al. Structural basis of kindlin-mediated integrin recognition and activation. Proc. Natl Acad. Sci. USA 114, 9349–9354 (2017).
Zhang, P. et al. Crystal structure of the FERM-folded talin head reveals the determinants for integrin binding. Proc. Natl Acad. Sci. USA 117, 32402–32412 (2020).
Legate, K. R. et al. Integrin adhesion and force coupling are independently regulated by localized PtdIns(4,5)2 synthesis. EMBO J. 30, 4539–4553 (2011).
Liu, J. et al. Structural basis of phosphoinositide binding to kindlin-2 protein pleckstrin homology domain in regulating integrin activation. J. Biol. Chem. 286, 43334–43342 (2011).
Qu, H. et al. Kindlin-2 regulates podocyte adhesion and fibronectin matrix deposition through interactions with phosphoinositides and integrins. J. Cell Sci. 124, 879–891 (2011).
Chinthalapudi, K., Rangarajan, E. S. & Izard, T. The interaction of talin with the cell membrane is essential for integrin activation and focal adhesion formation. Proc. Natl Acad. Sci. USA 115, 10339–10344 (2018).
Calderwood, D. A., Campbell, I. D. & Critchley, D. R. Talins and kindlins: partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 14, 503–517 (2013).
Anthis, N. J. et al. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 28, 3623–3632 (2009).
Wegener, K. L. et al. Structural basis of integrin activation by talin. Cell 128, 171–182 (2007).
Moser, M., Nieswandt, B., Ussar, S., Pozgajova, M. & Fassler, R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat. Med. 14, 325–330 (2008).
Bledzka, K. et al. Spatial coordination of kindlin-2 with talin head domain in interaction with integrin β cytoplasmic tails. J. Biol. Chem. 287, 24585–24594 (2012).
Fischer, L. S. et al. Quantitative single-protein imaging reveals molecular complex formation of integrin, talin, and kindlin during cell adhesion. Nat. Commun. 12, 919 (2021).
Yates, L. A., Fuzery, A. K., Bonet, R., Campbell, I. D. & Gilbert, R. J. Biophysical analysis of kindlin-3 reveals an elongated conformation and maps integrin binding to the membrane-distal β-subunit NPXY motif. J. Biol. Chem. 287, 37715–37731 (2012).
Kahner, B. N. et al. Kindlins, integrin activation and the regulation of talin recruitment to αIIbβ3. PLoS ONE 7, e34056 (2012).
Haydari, Z., Shams, H., Jahed, Z. & Mofrad, M. R. K. Kindlin assists talin to promote integrin activation. Biophys. J. 118, 1977–1991 (2020).
Anthis, N. J., Wegener, K. L., Critchley, D. R. & Campbell, I. D. Structural diversity in integrin/talin interactions. Structure 18, 1654–1666 (2010).
Jerabek-Willemsen, M. et al. MicroScale Thermophoresis: interaction analysis and beyond. J. Mol. Struct. 1077, 101–113 (2014).
Scheuermann, T. H., Padrick, S. B., Gardner, K. H. & Brautigam, C. A. On the acquisition and analysis of microscale thermophoresis data. Anal. Biochem. 496, 79–93 (2016).
Moore, D. T. et al. Affinity of talin-1 for the β3-integrin cytosolic domain is modulated by its phospholipid bilayer environment. Proc. Natl Acad. Sci. USA 109, 793–798 (2012).
Wishart, D. S. & Sykes, B. D. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4, 171–180 (1994).
Yang, J. et al. Structure of an integrin αIIbβ3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation. Proc. Natl Acad. Sci. USA 106, 17729–17734 (2009).
Helenius, J., Heisenberg, C. P., Gaub, H. E. & Muller, D. J. Single-cell force spectroscopy. J. Cell Sci. 121, 1785–1791 (2008).
Viljoen, A. et al. Force spectroscopy of single cells using atomic force microscopy. Nat. Rev. Methods Primers 1, 63 (2021).
Kondo, N., Ueda, Y. & Kinashi, T. Kindlin-3 disrupts an intersubunit association in the integrin LFA1 to trigger positive feedback activation by Rap1 and talin1. Sci. Signal. 14, eabf2184 (2021).
Orre, T. et al. Molecular motion and tridimensional nanoscale localization of kindlin control integrin activation in focal adhesions. Nat. Commun. 12, 3104 (2021).
Tan, S. J. et al. Regulation and dynamics of force transmission at individual cell–matrix adhesion bonds. Sci. Adv. 6, eaax0317 (2020).
Elliott, P. R. et al. The structure of the talin head reveals a novel extended conformation of the FERM domain. Structure 18, 1289–1299 (2010).
Bachir, A. I. et al. Integrin-associated complexes form hierarchically with variable stoichiometry in nascent adhesions. Curr. Biol. 24, 1845–1853 (2014).
Scholz, J. et al. A new method to customize protein expression vectors for fast, efficient and background free parallel cloning. BMC Biotechnol. 13, 12 (2013).
Tseng, H-Y. et al. LCP1 preferentially binds clasped αMβ2 integrin and attenuates leukocyte adhesion under flow. J. Cell Sci. 131, jcs218214 (2018).
Bottcher, R. T., Strohmeyer, N., Aretz, J. & Fassler, R. New insights into the phosphorylation of the threonine motif of the β1 integrin cytoplasmic domain. Life Sci. Alliance 5, e202101301 (2022).
Stark, H. GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 481, 109–126 (2010).
Klykov, O. et al. Efficient and robust proteome-wide approaches for cross-linking mass spectrometry. Nat. Protoc. 13, 2964–2990 (2018).
Perera, H. D. et al. Membrane binding of the N-terminal ubiquitin-like domain of kindlin-2 is crucial for its regulation of integrin activation. Structure 19, 1664–1671 (2011).
Lagerwaard, I. M., Albanese, P., Jankevics, A. & Scheltema, R. A. Xlink mapping and analysis (XMAS)—smooth integrative modeling in ChimeraX. Preprint at bioRxiv https://doi.org/10.1101/2022.04.21.489026 (2022).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Austen, K. et al. Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat. Cell Biol. 17, 1597–1606 (2015).
Meves, A., Stremmel, C., Bottcher, R. T. & Fassler, R. β1 integrins with individually disrupted cytoplasmic NPxY motifs are embryonic lethal but partially active in the epidermis. J. Invest. Dermatol. 133, 2722–2731 (2013).
Berginski, M. E. & Gomez, S. M. The Focal Adhesion Analysis Server: a web tool for analyzing focal adhesion dynamics. F1000Res. 2, 68 (2013).
Friedrichs, J., Helenius, J. & Muller, D. J. Quantifying cellular adhesion to extracellular matrix components by single-cell force spectroscopy. Nat. Protoc. 5, 1353–1361 (2010).
Yu, M., Strohmeyer, N., Wang, J., Müller, D. J. & Helenius, J. Increasing throughput of AFM-based single cell adhesion measurements through multisubstrate surfaces. Beilstein J. Nanotechnol. 6, 157–166 (2015).
Schubert, R. et al. Assay for characterizing the recovery of vertebrate cells for adhesion measurements by single-cell force spectroscopy. FEBS Lett. 588, 3639–3648 (2014).
Acknowledgements
We thank S. Bach for technical assistance, S. Uebel for help with MST measurements, β-CT synthesis and discussion of data, S. Suppmann, L. Urich and C. Strasser for protein expression, F. Brod and B. Steigenberger for help with cross-linking experiments, S. Asami and G. Gemmecker for NMR support and S. Grün for gel electrophoresis. This work was funded by the European Research Council (grant agreement no. 810104, point to R.F.), the DFG (SFB-863 to R.F. and SFB1035 and GRK1721 to M.S.), the Swiss National Science Foundation (grant no. 31003A_182587/1 to N.S.) and the Max Planck Society.
Funding
Open access funding provided by Max Planck Society.
Author information
Authors and Affiliations
Contributions
J.A. and R.F., conceptualization and writing original draft; J.A., M.A. and N.S., formal analysis, investigation, methodology and visualization; J.A., M.A., N.S. and M.S., writing (review and editing); M.S. and R.F., funding acquisition, resources and supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Sharon Campbell and Vesa Hytonen for their contribution to the peer review of this work. Primary Handling Editors: Sara Osman and Katarzyna Ciazynska, in collaboration with the Nature Structural & Molecular Biology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Analysis of ternary interactions of TLN1-F3, ΔKIND2 and β-CT.
a) Exemplary SDS-PAGEs of TLN1-F3, ΔKIND2, THD1, KIND2 and β1-TM-CT containing nanodisc preparations. b) Overlay of 1H-15N HSQC NMR spectra of 15N-labeled β1-CT before (black) and after addition of TLN1-F3 (green), ΔKIND2 (blue), or TLN1-F3 and ΔKIND2 (purple). Expansions of the overlaid spectra (i-iii) highlight the differences recorded in presence of TLN1-F3 or ΔKIND2 only, or TLN1-F3 and ΔKIND2 together. Arrows indicate changes of chemical shifts induced by addition of TLN1-F3 (green), ΔKIND2 (blue), or both proteins (purple). c) Overlay of methyl region of 1H-13C correlation NMR spectra of uniformly 13C,15N-labeled β1-CT before (black) and after addition of TLN1-F3 (blue), ΔKIND2 (green), or TLN1-F3 and ΔKIND2 together (purple). Methyl-containing amino acids observed in this experiment are highlighted in red in the β1-CT sequence (top). (i) highlights spectra shown in Fig. 1c and panel e. d) TLN1-F3 (200 µM; red) or ΔKIND2 (150 µM; orange) were preincubated with 1H-13C-labeled integrin β1-CT for 30 min at room temperature, then 150 µM ΔKIND2 was added to β1-CT/TLN1-F3 or 200 µM TLN1-F3 to β1-CT/ΔKIND2 samples and incubated for 10 min before NMR spectra were recorded. The methyl regions of the spectra are compared to those of β1-CT alone (black) and β1-CT incubated with 200 µM TLN1-F3 and 150 µM ΔKIND2 together (purple). Stepwise and simultaneous incubation of TLN1-F3, ΔKIND2 and β1-CT produced identical spectra indicating that ternary complexes form at equilibrium. e) Intensity ratio of peaks of the β1-CT in presence and absence of increasing concentrations of ΔKIND2 (top, green), TLN1-F3 (middle, blue), or both (bottom, magenta). Reported talin (blue) and kindlin (green) binding sites are indicated above the plots (MP: membrane proximal, MD: membrane-distal) and are reflected by line-broadening upon protein addition. The isolated peaks shown in Fig. 1b are indicated by (i) and (ii), respectively. Amino acid numbering is shown below plots. f) Exemplary MST traces of THD1 titrated to ATTO488-labeled β3-CT. To minimize heat effects, fluorescence changes were analyzed 1.5 s before (blue line) and after (red line) turning on the infrared laser. g) Titration of THD1 to ATTO488-labeled β1-CT in the presence (orange) or absence (blue) of 30 µM KIND2 (corresponding to 6-fold excess of Kd). h) Titration of KIND2 to ATTO488-labeled β1-CT in the presence (purple) or absence (green) of 300-400 µM THD1 (corresponding to 2-3-fold excess of Kd). Only concentrations up to about 10 µM KIND2 were considered and hence, non-linear regressions were extrapolated. Non-considered data points are shown in grey. i) Titration of THD2 to ATTO488-labeled β3-CT in the presence (orange) or absence (blue) of 30 µM KIND2 (corresponding to 3-fold excess of Kd). j) MST measurements of KIND2 affinity for ATTO488-labeled β3-CT in the absence (green) and presence of 250-450 µM TLN1-F3 (purple) or TLN2-F3 domain (dark purple; both corresponding to 3-5-fold excess of Kd). k) FC-RDA generated IC50 values of unlabeled THD1 competing binding of 565-THD1 (blue) and 647-KIND2 (orange), and unlabeled KIND2 competing binding of 647-KIND2 (green) and 565- THD1 (purple) to β1-TM-CT, β1-TM-CTY795A (KIND2-binding impaired), β1-TM-CTY783A (THD1-binding impaired), and α5-TM lacking α5-CT (α5-TM) embedded in 10% PIP2/90% PC-containing nanodiscs. Bars display mean ± SD (standard deviation) with a line for the median. l) MST affinity measurements of KIND2 for ATTO488-labeled talin-binding-impaired β1-CTY783A in absence (green) or presence (purple) of 400-600 µM THD1. m) MST affinity measurements of KIND2 for ATTO488-labeled talin-binding-impaired β3-CTY772A in absence (green) or presence (purple) of 500 µM THD1.
Extended Data Fig. 2 The α1-helix in TLN1-F3 binds to KIND2-F0.
a,b) 1H-15N HSQC NMR spectra of 15N-labeled TLN1-F3 (black) in the presence of KIND2 (a, green) or integrin β1-CT peptide (b, grey). c) CSP plot of 15N-labeled TLN1-F3 upon addition of 200 µM integrin β1-CT. Amino acid numbering is shown below the plot. d) CSPs recorded in 15N-labeled TLN1-F3 were mapped on crystal structure of TLN2-F3 (dark grey). e) 1H-15N HSQC NMR spectra of 15N-labeled TLN1-F3 in the presence of integrin β1-CT (black) and KIND2 (orange). f) CSP plot of 15N-labeled TLN1-F3 upon addition of 200 µM β1-CT together with 75 µM KIND2. Amino acid numbering is shown below the plot. g) CSPs recorded in uniformly 15N-labeled TLN1-F3 upon addition of β1-CT as well as KIND2 (indicated by orange spheres) mapped on the crystal structure of TLN2-F3. h) Top: Affinity reduction of THD1 mutants for β3-CT calculated by dividing the affinity of THD1 in absence (Kd,THD1) and presence of KIND2 (Kd,THD1+KIND2) measured by MST. Bottom: Bar chart of MST-derived affinities of indicated THD1 mutants for ATTO488- labeled β3-CT (n = 2) in absence (blue) and presence (orange) of 30-40 µM KIND2 (corresponding to 3-4-fold excess of Kd). i) Interprotein cross-links identified by mass spectrometry following disuccinimidyl suberate cross-linking of 60 µM TLN1-F3–β1-CT (top) and TLN1-F3K402E–β1-CT (bottom) fusion protein with 450 µM KIND2 using GraFix. j, k) 1H-15N HSQC NMR spectra of j) 100 µM 15N-labeled KIND2-F0 (black) in the presence of 100 µM TLN1-F3 (blue) or 100 µM TLN1-F3K402E (red) and of k) 100 µM 15N-labeled KIND2-F0Y13A (black) in the presence of 100 µM TLN1-F3 (light blue). Boxes indicate the region shown in Fig. 2h,i. l) CSP plot of 15N-labeled KIND2-F0 upon addition of 100 µM TLN1-F3 titration. Amino-acid numbering is shown below the plot. m) CSPs recorded in 15N-labeled KIND2-F0 were mapped on NMR structure of KIND2-F0 (blue). n) Peak intensity ratios in HSQC spectra of 15N-β1-CT in presence of increasing concentrations of ΔKIND2Y13A (yellow), TLN1-F3 and ΔKIND2Y13A (purple), and TLN1-F3K402E and ΔKIND2Y13A (pink). Talin (blue) and kindlin (green) binding sites in β1-CT are indicated above the plots (MP: membrane proximal, MD: membrane-distal). Stoichiometries of titrations are shown at the right side of plots and amino acid numbers below plots.
Extended Data Fig. 3 Characterization of talin-1K402E and kindlin-2Y13A mutants.
a,b) Similar 1H-15N HSQC NMR spectra of (a) 15N-labeled TLN1-F3 (blue) and TLN1-F3K402E (red) and (b) 15N-labeled KIND2-F0 (green) and KIND2-F0Y13A (yellow) confirm that the mutations do not perturb the overall fold of the proteins. c,d) CD spectra of (c) THD1 (blue) and THD1K402E (red) and (d) KIND2 (green) and KIND2Y13A (yellow) revealed identical secondary structures. e,f) Similar thermal stability of (e) THD1 (blue) and THD1K402E (red) and (f) KIND2 (green) and KIND2Y13A (yellow) determined by differential scanning fluorimetry. g,h) Representative dynamic light scattering measurement of (g) THD1 (blue) and THD1K402E (red) and (h) KIND2 (green) and KIND2Y13A (yellow), which excludes aggregate formation and hydrodynamic radius changes due to altered tertiary structure. Hydrodynamic radius obtained for THD1 was 3.4 ± 0.1 nm, for THD1K402E 3.2 ± 0.1 nm, and for KIND2 and KIND2Y13A 3.2 ± 0.1 nm (bar chart). i,j) SEC of (i) THD1 (blue) and THD1K402E (red) on Superdex75 and (j) KIND2 (green) and KIND2Y13A (yellow) on a Superdex200 excludes changes in hydrodynamic radius. Data are fitted to Gram-Charlier peak function resulting in identical maximum at 10.2 mL and a half-peak width of 0.34 mL for THD1 and THD1K402E and identical maximum at 13.1 mL and a half-peak width of 0.35 mL for KIND2 and KIND2Y13A. k) MST affinity measurements of KIND2 for ATTO488-labeled β3-CT in the absence (green) and presence (purple) of 120 µM THD1K402E (corresponding to 3-4-fold excess of Kd), and l) of KIND2Y13A for ATTO488-labeled β3-CT in absence (green) and presence (purple) of 300-400 µM THD1 (corresponding to 3-4-fold excess of Kd; n = 3). m) MST affinity measurements of THD1K402E for ATTO488-labeled β3-CT in the absence (blue) and presence (orange) of 30-40 µM KIND2 (corresponding to 3-4-fold excess of Kd), and n) of THD1 for ATTO488-labeled β3-CT in absence (blue) and presence (orange) of 30-40 µM KIND2Y13A (corresponding to 3-4- fold excess of Kd).
Extended Data Fig. 4 The talin and kindlin binding sites on β1-CT are allosterically coupled.
a) MST affinity measurements of THD1 for ATTO488- labeled β1-spacer-CT in absence (blue) and presence of 30-35 µM KIND2 (orange, corresponding to 6-7-fold excess of Kd), and b) of KIND2 for ATTO488-labeled β1-spacer-CT in absence (green) and presence of 300-400 µM THD1 (purple, corresponding to 2-3-fold excess of Kd).
Extended Data Fig. 5 Generation and characterization of TLN1K402E-YPet, mCherry-KIND2Y13A and β1-spacer expressing cells.
a) Cartoon depicting cell line generation. Upper image: mouse fibroblasts deficient of Talin-1, Talin-2, Fermt1 and Fermt2 (quadruple knockout; qKO) were reconstituted with mCherry-KIND2 or mCherry-KIND2Y13A together with either YPet-TLN1 (qKO-TLN1WTKIND2WT, black; qKO-TLN1WTKIND2Y13A, green) or YPet-TLN1K402E (qKO-TLN1K402EKIND2WT, blue; qKO-TLN1K402EKIND2Y13A, purple). Lower image: mouse fibroblasts deficient of Talin-1 and Talin-2 (double knockout, dKO) and expressing endogenous KIND2 were reconstituted with either YPet-TLN1 (dKO-TLN1WT; grey) or YPet-TLN1K402E (dKO-TLN1K402E; red). b,c) TLN1-YPet and mCherry-KIND2 levels in qKO cells (b) and surface integrin α5 and β1 in dKO cells (c) measured by flow cytometry. d) Quantification of TLN1-YPet, mCherry-KIND2, and integrin surface expression of indicated cells measured by flow cytometry. Data of qKO-TLN1K402EKIND2WT (blue), qKO-TLN1WTKIND2Y13A (green), qKO-TLN1K402EKIND2Y13A (purple), and dKO-TLN1K402E cells (red) was normalized to WT (black/grey) and analyzed for differences using a one sample t-test. e) Spreading area of indicated cells seeded on FN for indicated times. ≥50 cells (numbers indicated on top of each graph) were analyzed on three independent days. Statistics by one-way ANOVA for repeated measurements with Tukey post hoc test compared to dKO-TLN1WT or qKO-TLN1WTKIND2WT cells at identical timepoints). f) Spreading area of dKO-TLN1 cells seeded on FN-coated hydrogels with indicated Young’s modulus for 24 h. At least 50 cells were analyzed on three independent days. g) Scheme outlining the generation of mouse fibroblast cells deficient for β1 integrin (β1-KO, grey) reconstituted with human β1WT (blue) or β1-spacer (orange, Fig. 4c). h) Flow cytometry of β1WT and β1-spacer cells for 9EG7 levels normalized to total β1 integrin surface levels (n = 6). i) Plate-and-wash adhesion assays of serum-starved β1 knockout (KO), β1WT and β1-spacer cells 30 min after seeding on FN in absence and presence of MnCl2 and/or cilengitide (CGN). Data normalized to values from β1WT cells cultured in absence of MnCl2 and CGN (n = 3). j) Spreading area of serum-starved β1 knockout (KO), β1WT and β1-spacer cells 1 h after seeding on FN in absence and presence of MnCl2 and/or CGN. Cells were stained with DAPI and CellMask orange and areas of ≥50 cells were analyzed on three different days.
Extended Data Fig. 6 Overlay of THD1 structure corresponding to.
a) the canonical cloverleaf (dark blue; PDB: 6VGU) and b) an atypical linear conformation (light blue; 3IVF) with the model of TLN-F3-β1-CT-ΔKIND2 complex based on chemical crosslinks identified by mass spectrometry (Fig. 2e) using the reported structures of TLN2-F2F3 (blue)-β1D-CT (grey; PDB: 39 GW), ΔKIND2 (green)-β1A-CT (grey; PDB: 5XQ0) and KIND2-F0 (green; PDB: 2LGX). Note that the flexible N-terminus of KIND2-F0 occupies the space between the TLN1-F1 and TLN1-F3 domain in the cloverleaf fold (a) whereas the distance between the TLN1-F1 and TLN1-F3 domain suffice to accommodate the KIND2-F0 domain in the TLN1 linear fold (b).
Supplementary information
Supplementary Table 1
Chemical shift of KIND2-F0 transferred from BMRB 30659.
Supplementary Data 1
Exemplary gating of flow cytometry experiments with fibroblast cell lines.
Supplementary Table 2
Primers used in this study.
Supplementary Data 2
CSP analysis of NMR data.
Supplementary Data 3
Supporting data for Ca–Cb plots.
Source data
Source Data Figs. 1–5 and Extended Data Figs. 1–5
Data used to create all graphs in main and Extended Data figures.
Source Data Extended Data Fig. 1
Uncropped gel images of gels shown in Extended Data Fig. 1a.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Aretz, J., Aziz, M., Strohmeyer, N. et al. Talin and kindlin use integrin tail allostery and direct binding to activate integrins. Nat Struct Mol Biol 30, 1913–1924 (2023). https://doi.org/10.1038/s41594-023-01139-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-023-01139-9