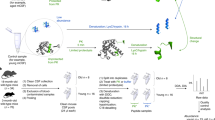
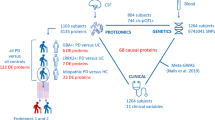
Abstract
Parkinson’s disease (PD) is a prevalent neurodegenerative disease for which robust biomarkers are needed. Because protein structure reflects function, we tested whether global, in situ analysis of protein structural changes provides insight into PD pathophysiology and could inform a new concept of structural disease biomarkers. Using limited proteolysis–mass spectrometry (LiP–MS), we identified 76 structurally altered proteins in cerebrospinal fluid (CSF) of individuals with PD relative to healthy donors. These proteins were enriched in processes misregulated in PD, and some proteins also showed structural changes in PD brain samples. CSF protein structural information outperformed abundance information in discriminating between healthy participants and those with PD and improved the discriminatory performance of CSF measures of the hallmark PD protein α-synuclein. We also present the first analysis of inter-individual variability of a structural proteome in healthy individuals, identifying biophysical features of variable protein regions. Although independent validation is needed, our data suggest that global analyses of the human structural proteome will guide the development of novel structural biomarkers of disease and enable hypothesis generation about underlying disease processes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The mass spectrometry proteomics dataset generated in this study is available in the PRIDE database111 (accession number PXD034120). Source data are provided with this paper.
Code availability
Code for the main analyses (Figs. 2b, 3, and 5) has been deposited on GitHub at https://github.com/beyergroup/Global-analyses-of-the-human-structural-proteome-to-identify-a-new-type-of-disease-biomarker. Further code for plots and other analyses is available upon request. Supplementary Table 12 contains all necessary data to use with the provided scripts.
References
Kennedy, B. K. et al. Geroscience: linking aging to chronic disease. Cell 159, 709–713 (2014).
Cilento, E. M. et al. Mass spectrometry: a platform for biomarker discovery and validation for Alzheimer’s and Parkinson’s diseases. J. Neurochem. 151, 397–416 (2019).
Crutchfield, C. A., Thomas, S. N., Sokoll, L. J. & Chan, D. W. Advances in mass spectrometry-based clinical biomarker discovery. Clin. Proteom. 13, 1 (2016).
Jiang, R. et al. Differential proteomic analysis of serum exosomes reveals alterations in progression of Parkinson disease. Medicine 98, e17478 (2019).
Macklin, A., Khan, S. & Kislinger, T. Recent advances in mass spectrometry based clinical proteomics: applications to cancer research. Clin. Proteomics 17, 17 (2020).
Thygesen, C., Boll, I., Finsen, B., Modzel, M. & Larsen, M. R. Characterizing disease-associated changes in post-translational modifications by mass spectrometry. Expert Rev. Proteom. 15, 245–258 (2018).
Tzeng, S. R. & Kalodimos, C. G. Protein activity regulation by conformational entropy. Nature 488, 236–240 (2012).
Henzler-Wildman, K. & Kern, D. Dynamic personalities of proteins. Nature 450, 964–972 (2007).
Schopper, S. et al. Measuring protein structural changes on a proteome-wide scale using limited proteolysis-coupled mass spectrometry. Nat. Protoc. 12, 2391–2410 (2017).
Feng, Y. et al. Global analysis of protein structural changes in complex proteomes. Nat. Biotechnol. 32, 1036–1044(2014).
Cappelletti, V. et al. Dynamic 3D proteomes reveal protein functional alterations at high resolution in situ. Cell 184, 545–559.e22 (2021).
Spillantini, M. G. et al. α-synuclein in Lewy bodies. Nature 388, 839–840 (1997).
Braak, H. et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 24, 197–211 (2003).
Brás, I. C., Xylaki, M. & Outeiro, T. F. Mechanisms of alpha-synuclein toxicity: an update and outlook. Prog. Brain. Res. 252, 91–129 (2020).
Maass, F., Schulz, I., Lingor, P., Mollenhauer, B. & Bähr, M. Cerebrospinal fluid biomarker for Parkinson’s disease: an overview. Mol. Cell. Neurosci. 97, 60–66 (2019).
Borrageiro, G., Haylett, W., Seedat, S., Kuivaniemi, H. & Bardien, S. A review of genome-wide transcriptomics studies in Parkinson’s disease. Eur. J. Neurosci. 47, 1–16 (2018).
Majbour, N. K. et al. Oligomeric and phosphorylated alpha-synuclein as potential CSF biomarkers for Parkinson’s disease. Mol. Neurodegener. 11, 7 (2016).
Parnetti, L. et al. CSF and blood biomarkers for Parkinson’s disease. Lancet Neurol. 18, 573–586 (2019).
van Dijk, K. D. et al. Changes in endolysosomal enzyme activities in cerebrospinal fluid of patients with Parkinson’s disease. Mov. Disord. 28, 747–754 (2013).
van Steenoven, I. et al. α-Synuclein species as potential cerebrospinal fluid biomarkers for dementia with lewy bodies. Mov. Disord. 33, 1724–1733 (2018).
Van Dijk, K. D. et al. Cerebrospinal fluid and plasma clusterin levels in Parkinson’s disease. Park. Relat. Disord. 19, 1079–1083 (2013).
van Dijk, K. D. et al. Reduced α-synuclein levels in cerebrospinal fluid in Parkinson’s disease are unrelated to clinical and imaging measures of disease severity. Eur. J. Neurol. 21, 388–394 (2014).
Abdi, I. Y. et al. Preanalytical stability of CSF total and oligomeric α-synuclein. Front. Aging Neurosci. 13, 85 (2021).
El‐Agnaf, O. M. A. et al. Detection of oligomeric forms of α‐synuclein protein in human plasma as a potential biomarker for Parkinson’s disease. FASEB J. 20, 419–425 (2006).
Oosterveld, L. P. et al. CSF biomarkers reflecting protein pathology and axonal degeneration are associated with memory, attentional, and executive functioning in early-stage Parkinson′s disease. Int. J. Mol. Sci. 21, 1–12 (2020).
Macron, C., Lane, L., Núnez Galindo, A. & Dayon, L. Deep dive on the proteome of human cerebrospinal fluid: a valuable data resource for biomarker discovery and missing protein identification. J. Proteome Res. 17, 4113–4126 (2018).
Barkovits et al. Blood contamination in CSF and its impact on quantitative analysis of α-synuclein. Cells 9, 370 (2020).
Macron, C. et al. Exploration of human cerebrospinal fluid: a large proteome dataset revealed by trapped ion mobility time-of-flight mass spectrometry. Data Brief 31, 105704 (2020).
Zhang, J. & Kurgan, L. SCRIBER: accurate and partner type-specific prediction of protein-binding residues from proteins sequences. Bioinformatics 35, i343–i353 (2019).
Jones, D. T. & Cozzetto, D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31, 857–863 (2015).
Beecham, G. W. et al. PARK10 is a major locus for sporadic neuropathologically confirmed Parkinson disease. Neurology 84, 972–980 (2015).
Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019).
Chang, D. et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 2017, 1511–1516 (2017).
Hoxha, E., Tempia, F., Lippiello, P. & Miniaci, M. C. Modulation, plasticity and pathophysiology of the parallel fiber-purkinje cell synapse. Front. Synaptic. Neurosci. 8, 35 (2016).
Lozovaya, N. et al. GABAergic inhibition in dual-transmission cholinergic and GABAergic striatal interneurons is abolished in Parkinson disease. Nat. Commun. 9, 1–14 (2018).
Zheng, X. et al. Increase in glutamatergic terminals in the striatum following dopamine depletion in a rat model of Parkinson’s disease. Neurochem. Res. 44, 1079–1089 (2019).
Gardoni, F., Ghiglieri, V., Luca, M. di & Calabresi, P. Assemblies of glutamate receptor subunits with post-synaptic density proteins and their alterations in Parkinson’s disease. Prog. Brain Res. 183, 169–182 (2010).
Błaszczyk, J. W. Parkinson’s disease and neurodegeneration: GABA-collapse hypothesis. Front. Neurosci. 10, 269 (2016).
Kayakabe, M. et al. Motor dysfunction in cerebellar Purkinje cell-specific vesicular GABA transporter knockout mice. Front. Cell. Neurosci. 7, 286 (2014).
Murueta-Goyena, A., Andikoetxea, A., Gómez-Esteban, J. C. & Gabilondo, I. Contribution of the GABAergic system to non-motor manifestations in premotor and early stages of Parkinson’s disease. Front. Pharmacol. 10, 1294 (2019).
Surmeier, D. J. et al. Calcium and Parkinson’s disease. Biochem. Biophys. Res. Commun. 483, 1013–1019 (2017).
Pchitskaya, E., Popugaeva, E. & Bezprozvanny, I. Calcium signaling and molecular mechanisms underlying neurodegenerative diseases. Cell Calcium 70, 87–94 (2018).
Latourelle, J. C. et al. Genomewide association study for onset age in Parkinson disease. BMC Med. Genet. 10, 98 (2009).
Blauwendraat, C. et al. Parkinson’s disease age at onset genome-wide association study: defining heritability, genetic loci, and α-synuclein mechanisms. Mov. Disord. 34, 866–875 (2019).
Tan, M. M. X. et al. Genome-wide association studies of cognitive and motor progression in Parkinson’s disease. Mov. Disord. 36, 424–433 (2021).
Wilhelmus, M. M. M. et al. Short communication apolipoprotein E and LRP1 increase early in Parkinson’s disease pathogenesis. Am. J. Pathol. 179, 2152–2156 (2011).
Troy T, R. & Jacob M, M. Apolipoprotein E fragmentation within lewy bodies of the human Parkinson’s disease brain. Int. J. Neurodegener. Disord. 1, 002 (2018).
Xu, J., Mashimo, T. & Südhof, T. C. Synaptotagmin-1, -2, and -9: Ca2+ sensors for fast release that specify distinct presynaptic properties in subsets of neurons. Neuron 54, 567–581 (2007).
Delignat-Lavaud, B. et al. The calcium sensor synaptotagmin-1 is critical for phasic axonal dopamine release in the striatum and mesencephalon, but is dispensable for basic motor behaviors in mice. Preprint at bioRxiv https://doi.org/10.1101/2021.09.15.460511 (2021).
Wu, M., Puddifoot, C. A., Taylor, P. & Joiner, W. J. Mechanisms of inhibition and potentiation of α4β2 nicotinic acetylcholine receptors by members of the Ly6 protein family. J. Biol. Chem. 290, 24509 (2015).
Wang, Q. et al. The landscape of multiscale transcriptomic networks and key regulators in Parkinson’s disease. Nat. Commun. 2019, 1–15 (2019). 101 10.
Power, J. H. T., Shannon, J. M., Blumbergs, P. C. & Gai, W. P. Nonselenium glutathione peroxidase in human brain: elevated levels in Parkinson’s disease and dementia with Lewy bodies. Am. J. Pathol. 161, 885–894 (2002).
Corradini, B. R. et al. Complex network-driven view of genomic mechanisms underlying Parkinson’s disease: Analyses in dorsal motor vagal nucleus, locus coeruleus, and substantia nigra. Biomed. Res. Int. 543673 (2014).
Lachén-Montes, M. et al. Unveiling the olfactory proteostatic disarrangement in Parkinson’s disease by proteome-wide profiling. Neurobiol. Aging 73, 123–134 (2019).
Lachén-Montes, M. et al. Smelling the dark proteome: functional characterization of PITH domain-containing protein 1 (C1orf128) in olfactory metabolism. J. Proteome Res. 19, 4826–4843 (2020).
Ma, S. et al. Peroxiredoxin 6 is a crucial factor in the initial step of mitochondrial clearance and is upstream of the PINK1–parkin pathway. Antioxid. Redox Signal. 24, 486–501 (2016).
Yun, H. M., Choi, D. Y., Oh, K. W. & Hong, J. T. PRDX6 exacerbates dopaminergic neurodegeneration in a MPTP mouse model of Parkinson’s disease. Mol. Neurobiol. 52, 422–431 (2015).
Elegheert, J. et al. Structural basis for integration of GluD receptors within synaptic organizer complexes. Science 353, 295–300 (2016).
Chipman, P. & Goda, Y in Dendrites: Development and Disease 425–465 (Springer Japan, 2016).
Won, S. Y., Lee, P. & Kim, H. M. Synaptic organizer: Slitrks and type IIa receptor protein tyrosine phosphatasess. Curr. Opin. Struct. Biol. 54, 95–103 (2019).
Lee, S. J. et al. Presynaptic neuronal pentraxin receptor organizes excitatory and inhibitory synapses. J. Neurosci. 37, 1062–1080 (2017).
Longhena, F., Faustini, G., Spillantini, M. G. & Bellucci, A. Living in promiscuity: the multiple partners of α-synuclein at the synapse in physiology and pathology. Int. J. Mol. Sci. 20, 141 (2019).
Fullard, M. E. & Duda, J. E. A review of the relationship between vitamin D and Parkinson disease symptoms. Front. Neurol. 11, 454 (2020).
Lawton, M. et al. Blood biomarkers with Parkinson’s disease clusters and prognosis: the oxford discovery cohort. Mov. Disord. 35, 279–287 (2020).
Li, T. & Le, W. Biomarkers for Parkinson’s disease: how good are they? Neurosci. Bull. 36, 183–194 (2020).
Kang, U. J. et al. Comparative study of cerebrospinal fluid α-synuclein seeding aggregation assays for diagnosis of Parkinson’s disease. Mov. Disord. 34, 536–544 (2019).
Rossi, M. et al. Ultrasensitive RT-QuIC assay with high sensitivity and specificity for Lewy body-associated synucleinopathies. Acta Neuropathol. 140, 49–62 (2020).
Rotunno, M. S. et al. Cerebrospinal fluid proteomics implicates the granin family in Parkinson’s disease. Sci. Rep. 2020, 1–11 (2020).
Eusebi, P. et al. Cerebrospinal fluid biomarkers for the diagnosis and prognosis of Parkinson’s disease: protocol for a systematic review and individual participant data meta-analysis. BMJ Open 7, e018177 (2017).
Simrén, J., Ashton, N. J., Blennow, K. & Zetterberg, H. An update on fluid biomarkers for neurodegenerative diseases: recent success and challenges ahead. Curr. Opin. Neurobiol. 61, 29–39 (2020).
Dixit, A., Mehta, R. & Singh, A. K. Proteomics in human Parkinson’s disease: present scenario and future directions. Cell. Mol. Neurobiol. 39, 901–915 (2019).
Parnetti, L. et al. Parkinson’s and Lewy body dementia CSF biomarkers. Clin. Chim. Acta 495, 318–325 (2019).
Heywood, W. E. et al. Identification of novel CSF biomarkers for neurodegeneration and their validation by a high-throughput multiplexed targeted proteomic assay. Mol. Neurodegener. 10, 64 (2015).
Magdalinou, N. K. et al. Identification of candidate cerebrospinal fluid biomarkers in parkinsonism using quantitative proteomics. Parkinsonism Relat. Disord. 37, 65–71 (2017).
Magdalinou, N., Lees, A. J. & Zetterberg, H. Cerebrospinal fluid biomarkers in parkinsonian conditions: an update and future directions. J. Neurol. Neurosurg. Psychiatry 85, 1065–1075 (2014).
Sarkar, A., Rawat, N., Sachan, N. & Singh, M. P. Unequivocal biomarker for Parkinson’s disease: a hunt that remains a pester. Neurotox. Res. 36, 627–644 (2019).
Liu, W. et al. Role of exosomes in central nervous system diseases. Front. Mol. Neurosci. 12, 240 (2019).
Postuma, R. B. et al. MDS clinical diagnostic criteria for Parkinson’s disease. Mov. Disord. 30, 1591–1601 (2015).
Geut, H. et al. Neuropathological correlates of parkinsonian disorders in a large Dutch autopsy series. Acta Neuropathol. Commun. 8, 39 (2020).
Hughes, A. J., Daniel, S. E., Kilford, L. & Lees, A. J. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: aclinico-pathological study of 100 cases. J. Neurol. Neurosurg. Psychiatry 55, 181–184 (1992).
Hoehn, M. M. & Yahr, M. D. Parkinsonism: onset, progression, and mortality. Neurology 17, 427–442 (1967).
Fahn, S. et al. The Unified Parkinson’s Disease Rating Scale. In Fahn, S. et al. (eds.) Recent Developments in Parkinson’s Disease, Vol. 2, 153-163 (1987).
Roth, M. et al. CAMDEX. A standardised instrument for the diagnosis of mental disorder in the elderly with special reference to the early detection of dementia. Br. J. Psychiatry 149, 698–709 (1986).
Alafuzoff, I. et al. Staging/typing of Lewy body related alpha-synuclein pathology: a study of the BrainNet Europe Consortium. Acta Neuropathol. 117, 635–652 (2009).
Thal, D. R. et al. Sequence of Aβ-protein deposition in the human medial temporal lobe. J. Neuropathol. Exp. Neurol. 59, 733–748 (2000).
Alafuzoff, I. et al. Staging of neurofibrillary pathology in Alzheimer’s disease: a study of the BrainNet Europe Consortium. Brain Pathol. 18, 484–496 (2008).
Mirra, S. S. et al. The consortium to establish a registry for Alzheimer’s disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 41, 479–486 (1991).
Montine, T. J. et al. National institute on aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol. 123, 1–11 (2012).
Thal, D. R., Griffin, W. S. T., de Vos, R. A. I. & Ghebremedhin, E. Cerebral amyloid angiopathy and its relationship to Alzheimer’s disease. Acta Neuropathol. 115, 599–609 (2008).
Kovacs, G. G. et al. Aging-related tau astrogliopathy (ARTAG): harmonized evaluation strategy. Acta Neuropathol. 131, 87–102 (2016).
Teunissen, C. E. et al. A consensus protocol for the standardization of cerebrospinal fluid collection and biobanking. Neurology 73, 1914–1922 (2009).
Bruderer, R. et al. Optimization of experimental parameters in data-independent mass spectrometry significantly increases depth and reproducibility of results. Mol. Cell. Proteom. 16, 2296–2309 (2017).
Reiter, L. et al. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry*. Mol. Cell. Proteom. 8, 2405–2417 (2009).
Savitski, M. M., Wilhelm, M., Hahne, H., Kuster, B. & Bantscheff, M. A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol. Cell. Proteom. 14, 2394–2404 (2015).
Muntel, J. et al. Surpassing 10,000 identified and quantified proteins in a single run by optimizing current LC–MS instrumentation and data analysis strategy. Mol. Omics 15, 348–360 (2019).
Bruderer, R. et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteom. 14, 1400–1410 (2015).
Teo, G. et al. MapDIA: preprocessing and statistical analysis of quantitative proteomics data from data independent acquisition mass spectrometry. J. Proteom. 129, 108–120 (2015).
Hui, W., Gel, Y. R. & Gastwirth, J. L. Lawstat: an R package for law, public policy and biostatistics. J. Stat. Softw. 28, 1–26 (2008).
Ameijeiras, J., Rosa, A., Crujeiras, M. & Rodríguez-Casal, A. multimode: an R package for mode assessment. J. Stat. Softw. 97, 1–32 (2018).
Buchan, D. W. A. & Jones, D. T. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 47, W402–W407 (2019).
Bonet, J., Harteveld, Z., Sesterhenn, F., Scheck, A. & Correia, B. E. Rstoolbox—a Python library for large-scale analysis of computational protein design data and structural bioinformatics. BMC Bioinform. 20, 240 (2019).
Zhao, B. et al. DescribePROT: database of amino acid-level protein structure and function predictions. Nucleic Acids Res. 49, D298–D308 (2021).
Peng, K., Radivojac, P., Vucetic, S., Dunker, A. K. & Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 7, 208 (2006).
Faraggi, E., Zhou, Y. & Kloczkowski, A. Accurate single-sequence prediction of solvent accessible surface area using local and global features. Proteins 82, 3170–3176 (2014).
Yan, J. & Kurgan, L. DRNApred, fast sequence-based method that accurately predicts and discriminates DNA-and RNA-binding residues. Nucleic Acids Res. 45, e84 (2017).
Szklarczyk, D. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Berman, H. M. et al. The protein data bank. Nucleic Acids Res. 28, 235–242 (2000).
Alexa, A. & Rahnenfuhrer, J. topGO: Enrichment analysis for gene ontology. Bioconductor R package (2020).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22 (2010).
Quast, J.-P., Schuster, D. & Picotti, P. protti: an R package for comprehensive data analysis of peptide- and protein-centric bottom-up proteomics data. Bioinform. Adv. 2, 1 (2022).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022).
Chen, H. M., Lin, C. Y. & Wang, V. Amyloid P component as a plasma marker for Parkinson’s disease identified by a proteomic approach. Clin. Biochem. 44, 377–385 (2011).
Dong, M. X. et al. Serum butyrylcholinesterase activity: A biomarker for Parkinson’s disease and related dementia. Biomed Res. Int. 2017, 1524107 (2017).
Acknowledgements
We gratefully acknowledge all individuals who donated samples used in this project. We thank: K. van Dijk and L. Oosterveld for help collecting CSF samples and clinical datasets; N. Majbour and O. El-Agnaf for collection of the alpha-synuclein datasets; and the Netherlands Brain Bank for postmortem brain tissue samples. M.-T. M. was supported by a long-term EMBO postdoctoral fellowship (ALTF 522-2019). L. N. was funded by DFG (grant CRC 1310 and grant agreement no. 398882498) and the German Academic Exchange Service (Forschungsstipendium fuer Doktorandinnen und Doktoranden). J. G. was funded by DFG (grants CRC 680 and CRC 1310). A. B. acknowledges funding by DFG (grant CRC 1310 and grant agreement no. 398882498). P. P. was funded by a Personalized Health and Related Technologies (PHRT) grant (PHRT-506), a Sinergia grant from the Swiss National Science Foundation (SNSF grant CRSII5_177195), the Peter Bockhoff Stiftung and the ETH Zurich Foundation, Parkinson Schweiz, the European Research Council (866004), and the EPIC-XS Consortium (823839), the last two under the EU Horizon 2020 program. W. D. J. v. d. B. was financially supported by grants from Amsterdam Neuroscience, Dutch Research council (ZonMW 70-73305-98-106; 70-73305-98-102; 40-46000-98-101), Michael J. Fox foundation (17253), and Dutch Parkinson Association (2020-G01). Some figures were created with BioRender.com.
Author information
Authors and Affiliations
Contributions
P. P. conceived the project. M.-T. M. conceived the experimental pipeline with input from P. P. and A. B. M.-T. M., A. B. and P. P. designed the experiments. M.-T. M. performed the experiments. M.-T. M., L. N. and F. S. analyzed the data. J. M., R. B., L. R. and W. D. J. v. d. B. collected the data. P. S. and M.-T. M. designed and analyzed in vitro experiments. W. D. J. v. d. B. provided the clinical samples. L. N. and J. G. performed the statistical analysis with input from A.B. N. d. S. supervised writing of the manuscript. M.-T. M., L. N., F. S., N. d. S., A. B. and P. P. wrote the manuscript. A. B. and P. P. supervised the project. All authors discussed and revised the final manuscript prior to submission.
Corresponding authors
Ethics declarations
Competing interests
The authors RB, JM and LR are full-time employees of Biognosys AG (Zurich, Switzerland). Spectronaut is a trademark of Biognosys AG. PP is an inventor of a patent licensed by Biognosys AG that covers the LiP–MS method used in this manuscript. WvdB performed contract research and consultancy for Hoffmann-La Roche, Roche Tissue Diagnostics, Crossbeta Sciences, Discoveric Bio and received research consumables from Hoffmann-La Roche and Prothena. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Structural and Molecular Biology thanks Tiago Outeiro, Marcus Bantscheff, and Laura Parkkinen for their contribution to the peer review of this work. Primary Handling editors: Anke Sparmann and Florian Ullrich, in collaboration with the Nature Structural & Molecular Biology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Global characteristics of study population.
a, Characteristics of the study cohort. P values were estimated via Wilcoxon rank sum test. b, Age distribution within the healthy (HG) and PD (PDG) cohort groups, separated by sex. Boxplots: median, center; first and third quantile, lower and upper hinges; largest/smallest value no further than 1.5 * inter-quantile range of the hinge, whiskers; data points beyond are defined as outliers and plotted individually. P values are indicated (Wilcoxon rank sum test, n = 51 subjects in HG, n = 52 subjects PDG). c, GO enrichment of all identified proteins in the CSF proteome (trypsin-only control data) using the human proteome (UniProt FASTA, July 2019) as the background. Only the 10 terms with the highest enrichment per GO domain are shown. Numerical data for graphs in b and c are available as source data.
Extended Data Fig. 2 Comparison of structural features of variable and non-variable peptides, and the proteins containing these peptides, in CSF.
a, Distribution of secondary structures as loops, alpha-helix and beta-strands for variable/non-variable peptides, as predicted using PSIPRED. Boxplots for all panels: median, center; first and third quantile, lower and upper hinges; largest/smallest value no further than 1.5 * inter-quantile range of the hinge, whiskers; data points beyond are defined as outliers and plotted individually. P values are indicated (Wilcoxon rank sum test; ns, not significant; 9385 non-variable, 386 medium-variable, 117 high-variable peptides, from 51 subjects) b, c, Predicted propensity of peptides to bind DNA or RNA. d, Predicted solvent accessible surface area of variable/non-variable peptides. e, Number of high confidence interaction in STRING for proteins with at least one highly variable peptide (red), as compared to all other proteins (gray). f, g, Sequence length and number of domains as annotated in the PFAM database for proteins with at least one highly variable peptide (red), as compared to all other proteins (gray). h, Side-by-side view of affected peptides from in situ (left, reproduced from Fig. 2g for comparison) and in vitro experiments (right). Structure of human brain fructose bisphosphate aldolase (PDB entry 1XFB). The enzyme is a homotetramer, one subunit is shown as light blue cartoon and the other 3 subunits are shown as gray surface. The substrate is represented as yellow spheres, based on an alignment of PDB entry 1XFB with the ligand bound structure of the muscle isoform (PDB entry 4ALD). For the in situ data (left), the highly variable peptides are highlighted in dark red (bimodal) and salmon (unimodal). For the in vitro data (right), the significant peptides in the presence and absence of fructose 1,6-bisphosphate are highlighted in red (log2 fold change < −1 or > 1). The bimodal and non-bimodal peptide identified in situ are encircled. Numerical data for graphs in a-g are available as source data.
Extended Data Fig. 3 Effects of the sex variable on the linear model and overlap between the brain and CSF data sets.
a, The histogram visualizes the P values (calculated via t-statistics) of the cohort variable estimated from the linear model describing effects of structural variation, with the indicated combinations of the sex variable and interactions with sex taken into account. For all models, the first bar (extreme left) indicates significant (<0.05) P values. b, Number of proteins and peptides of the CSF and brain samples used for estimating the linear models, visualizing the overlapping peptides and proteins between the two tissues. c, Number of candidate peptide from the CSF where the coefficients change in the same or different direction in the brain samples. Numerical data for graphs in a and c are available as source data.
Extended Data Fig. 4 Structural changes in selected CSF and brain proteins.
a, b, Structure of PSAT1 (PDB 3E77) colored according to peptides in CSF (a) and brain (b) data. PSAT catalyzes an important step in serine biosynthesis. Black indicates all analyzed peptides; red indicates the candidate peptide (a) or peptides with a significant P value (b). One candidate CSF peptide is only 6.0 Å away from the active site. c, Coverage plot for all analyzed PSAT1 peptides in CSF (top) and brain (bottom). Black represents fully tryptic; gray represents half tryptic peptides and red bars the candidate/significant peptide in the CSF/ brain. d, e, Structure of PRDX6 (PDB PRX1) colored according to peptides in CSF (d) and brain (e) data. Colors as in a, b. f, Coverage plot for all analyzed PRDX6 peptides in CSF (top) and brain (bottom). Colors as in c. g, Structure of AFM (PDB 5OKL). h, Coverage plot for AFM. AFM level in serum exosomes was linked to PD progression4. i, Scaled residual plots for AFM. Boxplots are as in Extended Data Fig. 2 (n = 51 subjects in each HG and PDG). j, Structure of SAMP (PDB entry 1GYK) as pentamer with bound calcium (yellow spheres), an abundance-based plasma biomarker candidate for PD112. k, Coverage plot of SAMP. l, Scaled residual plots for SAMP (ERVGEYSLYIGR: n = 47 subjects in HG and n = 50 subjects in PDG; IVLGQEQDSYGGK: n = 51 subjects in each HG and PDG). m, Structure of BCHE (PDB entry 1P0I) in complex with butanoic acid (yellow spheres). Activity of this enzyme is decreased in PD with dementia (PDD)113, note that BCH inhibition was not used in our cohort. n, Coverage plot for BCHE. o, Scaled residual plot for BCHE (NIAAFGGNPK: n = 51 subjects in each HG and PDG; IFFPQVSEFGK: n = 43 subjects in HG and n = 48 subjects in PDG). Peptide colors in panels G, J, M are as in D; colors in panels H, K, N are as in C. Numerical data for graphs in c, f, h, i, k, l, n and o are available as source data.
Extended Data Fig. 5 Classification of Parkinson’s Disease in CSF data.
a, ROC curves for classification of PD based on LiP peptide variation. In this case, LiP peptide intensities were neither corrected for trypsin-only peptide intensities nor for protein abundance. b, ROC curves for classification of PD based on LiP peptide variation. In this case, LiP peptide intensities were not corrected for protein abundance. c, ROC curves for classification of PD based on LiP peptide variation. In this case, LiP peptide intensities were not corrected for trypsin-only peptide intensities. d, ROC curves for classification of PD based on ELISA measurement of different a-synuclein species from. e, Total α-synuclein levels compared to the ratio of the oligomeric/total α-synuclein level across the cohort. f, Oligomeric α-synuclein levels compared to the ratio of the oligomeric/total α-synuclein level across the cohort. For (e) and (f), each dot represents a single individual. g, Comparison of classification of the PDG using the ratio of oligomeric to total a-synuclein (log odds plotted on x axis) and using a combination of five LiP peptide levels (log odds plotted on y axis). Each point represents an individual and the HY-stage is indicated by color. Numerical data for graphs in a-g are available as source data.
Supplementary information
Supplementary Table 1
Raw peptide intensities of healthy and PD CSF samples measured for trypsin-only (PK-independent) conditions.
Supplementary Table 2
Raw peptide intensities of healthy and PD CSF samples measured for LiP conditions.
Supplementary Table 3
Summary of the number of peptides and proteins used in different analyses of CSF and brain data.
Supplementary Table 4
Protein and peptide usage across analyses of the CSF samples.
Supplementary Table 5
Structural peptide variability analysis. Peptides included in the analysis of the variability of the healthy CSF human proteome.
Supplementary Table 6
LiP–MS analysis of in vitro FBP aldolase and vitamin-D-binding protein with and without substrate.
Supplementary Table 7
Structural variation in PD model results.
Supplementary Table 8
PK-independent variation in PD model results.
Supplementary Table 9
Protein abundance variation in PD model results.
Supplementary Table 10
Overview of all structurally changed candidate peptides in CSF.
Supplementary Table 11
Overlap of candidate CSF biomarkers with brain data. List of CSF candidate biomarkers for structural rearrangements in PD which also show a structural change in the brain data.
Supplementary Table 12
Preprocessed data for code deposited on GitHub.
Source data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Statistical Source Data
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mackmull, MT., Nagel, L., Sesterhenn, F. et al. Global, in situ analysis of the structural proteome in individuals with Parkinson’s disease to identify a new class of biomarker. Nat Struct Mol Biol 29, 978–989 (2022). https://doi.org/10.1038/s41594-022-00837-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-022-00837-0