Abstract
Drug discovery campaigns against COVID-19 are beginning to target the SARS-CoV-2 RNA genome. The highly conserved frameshift stimulation element (FSE), required for balanced expression of viral proteins, is a particularly attractive SARS-CoV-2 RNA target. Here we present a 6.9 Å resolution cryo-EM structure of the FSE (88 nucleotides, ~28 kDa), validated through an RNA nanostructure tagging method. The tertiary structure presents a topologically complex fold in which the 5′ end is threaded through a ring formed inside a three-stem pseudoknot. Guided by this structure, we develop antisense oligonucleotides that impair FSE function in frameshifting assays and knock down SARS-CoV-2 virus replication in A549-ACE2 cells at 100 nM concentration.
Similar content being viewed by others
Main
Since December 2019, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus, the etiological agent of coronavirus disease 2019 (COVID-19), has been a pandemic-scale, global health concern. As of February 2021, more than 103 million people have been infected and more than 2.2 million have died, and this number is still growing. Effective vaccines and therapeutic drugs are needed to reduce the severity of the ongoing SARS-CoV-2 global pandemic, particularly as novel viral variants emerge that may be more infectious and/or escape current vaccination efforts. As of early 2021, 183 vaccines and ~446 therapeutic drugs are being developed worldwide for combating SARS-CoV-2 infection, although most are focused on a few targets1. For other diseases, specific drug design based on protein and RNA structures has been an effective means of drug development2,3,4. Three-dimensional (3D) structures of several key proteins of SARS-CoV-2 have been resolved5,6, but 3D structures of its key RNA elements remain structures of its key RNA elements remain poorly characterized.
Like all coronaviruses, SARS-CoV-2 has a positive-sense (+), single-stranded RNA (ssRNA) genome. Its first open reading frames (ORFs) 1a and 1b, which partially overlap, encode essential SARS-CoV-2 non-structural proteins, including the RNA-dependent RNA polymerase7. Optimal viral fitness is dependent on precise stoichiometric expression of ORF1a and ORF1ab throughout the viral replication cycle7,8, facilitated by an essential regulatory mechanism termed −1 programmed ribosomal frameshifting (−1 PRF). The −1 PRF mechanism is stimulated by a structured RNA motif at the 3′ end of ORF1a termed the frameshift stimulation element (FSE). This element directs elongating ribosomes to stochastically shift their reading frames by one base in the 5′ direction, enabling readthrough past the ORF1a stop codon into ORF1b and maintaining appropriate ratios of ORF1a to ORF1ab expression9.
SARS-CoV-2 has an FSE sequence identical to the original SARS-CoV-1 FSE up to a single-nucleotide substitution10 (non-synonymous mutation C13533A). As with other coronavirus FSEs, the SARS-CoV-2 FSE has a 5′ heptanucleotide ‘slippery site’ UUUAAAC followed by an RNA element hypothesized to form a three-stem pseudoknot9,11. Sequence conservation, in vitro functional analysis and small-angle X-ray scattering (SAXS) suggest that the SARS-CoV-2 forms an FSE that folds similarly to the original SARS coronavirus FSE. The SARS-CoV-2 FSE is highly conserved compared to its genomic context, with few single-nucleotide polymorphisms arising in recent strains, all of which sustain frameshifting in in vitro assays12. Consequently, the SARS-CoV-2 FSE appears functionally obligate for viral fitness and is predicted to have little evolutionary flexibility to evolve away from any potential therapeutics13. Supporting its potential as a therapeutic target, previous studies have shown that changing the FSE sequence leads to loss of viral fitness in SARS-CoV-1 and other coronaviruses7,8. Secondary structures required for coronavirus frameshifting have been determined by NMR14, compensatory mutagenesis13 and nucleotide-resolution chemical mapping, and have informed extensive genetic, biochemical and biophysical studies13,15. Attempts to disrupt coronavirus frameshifting have validated the FSE as a promising target, but the resulting peptide nucleic acids, locked nucleic acids (LNAs) and small molecules have required greater than micromolar concentrations to inhibit SARS-CoV-2 in cells and have not yet resulted in usable therapeutics16,17,18.
In this Article, we report structural and functional results relevant for efforts to disrupt SARS-CoV-2 through FSE targeting. In an effort to facilitate structure-guided drug design, we report a 3D structure of an 88-nucleotide SARS-CoV-2 FSE RNA at 6.9-Å resolution by single-particle cryo-EM and validate it by a second cryo-EM analysis involving a rationally designed RNA nanostructure. The structure shows a tertiary arrangement in which the 5′ end is threaded through a ring formed inside a three-stem pseudoknot and suggests mechanistic insights into how frameshifting might be inhibited efficiently. Antisense oligonucleotides (ASOs) informed by these insights give dose-dependent inhibition of −1 PRF in in vitro frameshifting assays and of SARS-CoV-2 replication in A549-ACE2 cells. These results demonstrate the therapeutic potential of targeting the SARS-CoV-2 frameshift stimulation element, establish ASO leads for pre-clinical testing, and provide a structural and functional framework for further development of both ASO and small-molecule compounds.
Results
Cryo-electron microscopy resolves the 88-nucleotide FSE
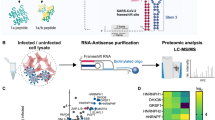
Cryo-EM structure determination of RNA-only systems or macromolecules with molecular weights under 50 kDa has been accomplished only recently19,20,21,22,23. At the onset of our study, it was unclear whether cryo-EM would be applicable to an RNA as small as the SARS-CoV-2 FSE (28 kDa, 88 nucleotides) (Fig. 1a). Initially, we collected a dataset comprising ~13,000 micrographs. However, the low concentration of the sample on the grids and the high background of molecular species in the raw images made it difficult to perform image processing (Extended Data Fig. 1). We manually selected ~3,400 images out of the whole dataset with defocus higher than −3 μm, with the aim of achieving an initial 3D reconstruction from ~27,000 manually picked particles (Extended Data Fig. 1). Subsequently, we optimized the sample preparation and RNA folding protocol to improve the stability and concentration of stably folded particles on the electron microscope grids (Methods). A dataset with ~10,000 micrographs was collected using the optimized sample (Supplementary Table 1). Benefiting from the lower background, about one million particles were selected with the ‘Neural Network’ particle autopicking option in EMAN224 and were subject to 2D and 3D image classification in Relion25 (Extended Data Fig. 1 and Supplementary Fig. 1). We selected one class of images yielding a reconstruction with well-connected density for further refinement. The reference-free 2D classification of this particle set and its refined 3D map resemble the Greek character ‘λ’ (Fig. 1 and Extended Data Fig. 1) at a final resolution of 6.9 Å (Fig. 1b). The B factor26,27, calculated based on a fitted relationship of particle number and map resolution, was estimated to be ~726 Å2 (Supplementary Fig. 2). Such a large B factor could be attributable to the flexibility of the molecule and/or the limit of particle orientation estimation accuracy due to its small size. Nevertheless, at the current 6.9 Å resolution, the map still permitted identification of the helical groove features that are hallmarks of RNA (Fig. 1c). The map also showed non-helical crevices and holes suggestive of potential small-molecule binding pockets (Fig. 1c), as previously observed and validated in cryo-EM maps of S-adenosylmethionine-IV (SAM-IV) riboswitch aptamers, with and without small-molecule ligands21,23.
a, Reference-free 2D class averages. b, Gold standard Fourier shell correlation plots calculated in Relion. c, Reconstructed cryo-EM map in three different views. d, Secondary structure of the FSE as determined by 1D SHAPE chemical mapping. e, The top 10 tertiary structures of the FSE as determined by auto-DRRAFTER using the secondary structure from d, where equivalent structural elements are indicated. These 10 top scoring models had a mean pairwise r.m.s.d. of 5.68 Å, resulting in an estimated accuracy of 5.9 Å based on the previously determined linear relationship23. f, The first strand of stem 1 is captured within a ring formed by the second strand of stem 1 with stems 2 and 3 and J3/2, producing a topologically constrained tertiary fold.
Ribosolve modeling of FSE coordinates
To better understand the FSE tertiary structure, we sought to model RNA coordinates into the 6.9-Å map using Ribosolve, a hybrid pipeline recently developed for automatically modeling RNA 3D structures based on secondary structure information from mutate-and-map guided by next-generation sequencing (M2-seq), cryo-EM maps and computer modeling with auto-DRRAFTER23. M2-seq secondary structure analysis recovered the three-stem pseudoknot in Fig. 1a, which has been validated by NMR14 and compensatory mutagenesis for the SARS-CoV-1 FSE13. The same three-stem secondary structure was also observed in a separate SHAPE-directed modeling study as well as independent DMS-MaPseq and SHAPE-MaPseq studies, with minor variations in edge base pairs (Extended Data Fig. 2). Using the consensus secondary structure of Fig. 1d and the cryo-EM map, auto-DRRAFTER modeling produced an ensemble of 10 structures with a mean pairwise root mean squared deviation (r.m.s.d.) of 5.68 Å (Fig. 1e, Extended Data Fig. 3 and Supplementary Video 1), corresponding to a 5.9-Å estimated accuracy, based on a linear calibration of precision and accuracy performed previously23. The quality of the resulting model ensembles was validated using MolProbity28 (Table 1). At an estimated accuracy of 5.9 Å, individual atomic positions and non-canonical base pairs cannot be assigned confidently. Nevertheless, the tertiary arrangement of the helical segments and non-helical linkers of the SARS-CoV-2 FSE can be traced (Fig. 1). This 3D architecture is consistent in different members of the model ensemble and is further supported by alternate auto-DRRAFTER runs making different assumptions about secondary structure and how much of the 5′ end is resolved in the map (Extended Data Fig. 3).
Overall architecture of the SARS-CoV-2 FSE
The architecture of the FSE involves several interlocking elements (Fig. 1e,f and Supplementary Video 1). Starting from the 5′ end and proceeding to the 3′ end, the molecule begins with a 5′ region that includes the heptanucleotide slippery site. The Ribosolve modeling folds this region into a loose hairpin-like shape closed by G•U wobble pairs, although the sparse cryo-EM map density in the region and chemical mapping is also consistent with a partially structured single-strand conformation (Extended Data Fig. 3d,e). This 5′ end is followed by the first strand of stem 1, a long helix in all coronavirus FSEs. The loop of stem 1 is also the first strand of the stem 2 pseudoknot, which forms a 5-bp helix that hybridizes with its complement at the 3′ end of the FSE. The RNA strand continues from this region to complete the second strand of stem 1 and doubles back to form a hairpin, stem 3. After an unpaired segment J3/2, the RNA completes stem 2 to close the stem 1-stem 2 pseudoknot. Finally, unstructured terminal nucleotides, invisible to cryo-EM, form a 3′ tail.
Despite the absence of base pairings or direct stacking between stem 3 and the stem 1-stem 2 pseudoknot, stem 3 exhibits a distinct tertiary conformation in relation to the pseudoknot, which, along with the conformationally heterogeneous 5′ end, result in the legs of the ‘λ’-shaped map (Fig. 1e,f, Extended Data Fig. 4 and Supplementary Video 1). Overall, stems 1, 2 and 3 form a circular ring with a visually apparent hole (Fig. 1c–f). The 5′ end of the FSE is connected to the pseudoknot by a linker that is threaded through this ring. This complex topology was predicted as a possible FSE fold in two recent, independent 3D computer modeling studies that include an extended 5′ end. We note that, although the fold of the 5′ end may have multiple conformations, the ring-threaded 5′ end is a consistent feature in all models in the auto-DRRAFTER ensemble (Supplementary Video 1) as well as in alternate modeling runs based on alternate FSE secondary structures and different auto-DRRAFTER modeling assumptions (Extended Data Fig. 3). In terms of structural requirements, the capture of the 5′ end in the stem 1–stem 2–stem 3 ring requires formation of a ~10-bp helical turn in stem 1. With fewer base pairs, the 5′ strand of stem 1 cannot turn fully inside and through the ring (Supplementary Fig. 7). Supporting the general relevance of the ring-threaded 5′ end, the length of stem 1 is ~10 bp or larger in all proposed coronavirus FSE elements, and de novo computer modeling of FSEs from murine hepatitis virus (MHV), human coronaviruses HKU1 and OC43, bovine coronavirus (BCoV), Middle Eastern respiratory syndrome-related coronavirus (MERS), SARS-CoV-1 and SARS-CoV-2 all give models with ring-threaded 5′ ends (Extended Data Fig. 5).
Validation of the FSE model by RNA nanostructure tagging
The SARS-CoV-2 FSE RNA represents an extreme case for cryo-EM. It is one of the smallest macromolecules (28 kDa) so far resolved by cryo-EM single-particle analysis at subnanometer resolution. We therefore sought further independent validation of the map and the Ribosolve model, particularly to test our inference that stem 3 comprises a ‘leg’ of the λ shape, perpendicular to, rather than coaxially stacked to, the stem 1-stem 2 pseudoknot. We rationally designed a variant of the SARS-CoV-2 FSE termed FSE-ATP-TTR3, which contains an insertion of a clothespin-like nanostructure whose visualization would test the assignment and orientation of stem 3 in the FSE map29 (Fig. 2a). For the tag, we chose the rationally designed ATP-TTR 3 RNA30, based on its known amenability to cryo-EM imaging23 and on modeling suggesting that its insertion into the FSE would not perturb either RNAs’ secondary structure, a prediction verified through chemical mapping (Extended Data Fig. 6).
a, Secondary structure of the FSE-ATP-TTR3 as determined by SHAPE chemical mapping, with key structural elements indicated in different colors. b, Cryo-EM map of the FSE-ATP-TTR3 at a low contour level, oriented as in a. c, ATP-TTR 3 model (PDB ID 6WLK) docked into the FSE-ATP-TTR3 map. d, FSE map from Fig. 1c, green, docked into the extra density of the FSE-ATP-TTR3 map. e, Comparison between the FSE map and the extra density of the FSE-ATP-TTR3 map.
A dataset containing ~12,000 micrographs of FSE-ATP-TTR3 was collected and resulted in a 6.4-Å map of FSE-ATP-TTR3 that clearly contained the ATP-TTR 3 clothespin-like shape. Additional density at the end of the clothespin is visible, with a λ-like shape (Fig. 2b,c and Extended Data Fig. 7). The FSE-only map fits into this additional density (Fig. 2d). To evaluate the relative orientation of FSE and the ATP-TTR tag, we adopted an unbiased method to align the two maps. The orientation of the FSE map was determined by conducting a rigid-body exhaustive search to maximize the correlation between the FSE density and difference mapping between FSE-ATP-TTR3 and ATP-TTR3 densities independently using Segger31 and Situs32. The highest cross-correlation was found at the expected site, supporting the orientation and location of stem 3 within the FSE map (orange helix, Fig. 1e; Supplementary Fig. 4) and prospectively corroborating the prediction from auto-DRRAFTER analysis of the FSE alone.
Targeting of SARS-CoV-2 FSE using LNA
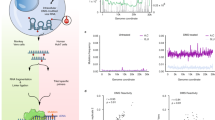
Several studies have proposed the FSE as a target for disruption of virus replication but have not previously yielded antiviral compounds effective at the submicromolar concentrations that might lead to effective therapies7,8. Before our cryo-EM results, we designed a first generation of LNA ASOs targeting FSE stems 2 and 3, shown in Fig. 3a. We initially avoided targeting stem 1 because of its perceived inaccessibility to antisense hybridization in the secondary structure, as well as unfavorable predicted properties of GC-rich LNAs needed to target such regions33,34,35. Cell-free in vitro dual-luciferase frameshifting assays confirmed the ability of these ASOs to reduce frameshifting, although the inhibition was partial (two- to fivefold at saturating concentrations) (Methods and Fig. 3b) (Extended Data Fig. 8a). The ASOs were further tested for antiviral potential in Huh-7 and Vero-E6 cell lines against a fully replicating SARS-CoV-2 luciferase reporter virus (SARS-CoV-2-nLuc) (Fig. 3c,d). At 25 nM (chosen to remain well below the cytotoxicity thresholds of ~1 μM) (Extended Data Fig. 9), the ASOs achieved observable inhibition compared to a Scramble LNA negative control. However, the overall effect was less than fourfold (<0.6 log10 inhibition) in both cell lines, compared to 2.3 log10 inhibition for the positive control compound, EIDD-1931 (NHC, β-d-N4-hydroxycytidine, a ribonucleoside analog)36.
a, ASOs modified with LNA bases that were designed against the SARS-CoV-2 FSE RNA secondary structure (colored lines). Ambiguous base-pairing partners are indicated with dashed lines. The C13533A mutation between SARS-CoV-1 and SARS-CoV-2 is shown in red. b, Mean in vitro dual luciferase frameshifting assay of initial, pre-structure informed LNAs, as a function of LNA challenge, as shown as percent frameshifting ± standard deviation. Values are normalized as described in the Methods. Fits are derived from a standard binding isotherm fit to the data (Methods) to estimate IC50 values versus SARS-CoV-2 (±standard error in stated units; Methods): S2D 1.9 ± 1.6 µM; S3D-1 500 ± 90 nM; S3D-2 180 ± 20 nM. c,d, FSE-LNA inhibition of SARS-CoV-2-nLuc virus replication in Huh-7 (c) and Vero E6 (d). Cells were treated with 25 nM of FSE-directed or Scrambled (Scr) LNAs 24 h before infection with SARS-CoV-2-nLuc reporter virus. At 48 h post-infection, luciferase expression was measured. The nucleoside analog, EIDD The nucleoside analog, EIDD-1931 (EIDD), was used as a positive control. Results are shown as log10 luciferase expression; n = 2. e, Mean in vitro dual luciferase frameshifting assay of initial, post-structure informed LNAs, as a function of LNA challenge, as shown as percent frameshifting ± standard deviation. Fits are derived as in b. Slp2 320 ± 40 nM; S1D-3 280 ± 90 nM; S1D-1 130 ± 20 nM; S1D-2 280 ± 70 nM. f, FSE-LNA inhibition of SARS-CoV-2-nLuc virus replication in ACE2-A549 cells. LNA treatment and infection were performed as described in c and d with 25 nM and 100 nM LNA concentrations. Results are shown as log10 luciferase expression, n = 4. P values were generated by GraphPad Prism software and computed as an ordinary one-way analysis of variance (ANOVA) using Dunnett’s multiple comparisons test against the Scr. LNA mean. Error bars represent ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. NS, not significant.
After our cryo-EM structure, we revisited stem 1 as a target for ASO hybridization. The 5′-end threaded structure (Fig. 1f) suggested a potential ‘torsional restraint’ model37 for frameshift stimulation by the SARS-CoV-2 FSE (Discussion), and thereby implicated stem 1 with a central structural role in −1 PRF. Furthermore, the tertiary structure showed similar accessibilities of stem 1 as with stem 2 and stem 3, our previous targets. These observations inspired us to design ASOs that might directly disrupt some or all of stem 1 after gaining toeholds in the nearby single-stranded regions, despite the aforementioned expected difficulties with targeting the region. In vitro frameshifting assays of the new stem1-targeted ASOs demonstrated greater inhibitory action (Fig. 3e) compared to the original stem 2 and stem 3-targeted LNAs (Fig. 3b). Encouraged by these results, the ASOs were tested against SARS-CoV-2-nLuc. These experiments were carried out in a newly developed ACE2-A549 cell line38, designed to better reproduce and replicate SARS-CoV-2 infection. Cytotoxicity assays also indicated that the new LNAs could be tolerated at higher concentrations than the first generation, motivating tests at both 25 nM and 100 nM ASO doses (Extended Data Fig. 9). In contrast to the modest results of stem 2 and 3 disrupting ASOs (Fig. 3c,d), stem 1 and slippery site-targeting ASOs displayed greater activity against SARS-CoV-2 replication, with S1D-2 giving 1.6 log10 inhibition compared to a scramble LNA negative control, and even outperforming the positive control EIDD-1931 by 0.8 log10 (Fig. 3f). Furthermore, the dose dependence (ASO effects at 25–100 nM) (Fig. 3f) and a correlation of in vitro frameshifting activity versus inhibition in cells (r2 = 0.44, P < 0.0025) (Extended Data Fig. 10) supported a mechanism of virus inhibition involving hybridization to the FSE and disruption of frameshifting.
Discussion
The urgency of the COVID-19 pandemic, recent advances in targeting RNA structures with ASOs and small molecules, and the identification of the FSE as a potentially well-defined RNA 3D structure in the SARS-CoV-2 genome has generated strong interest in understanding and targeting SARS-CoV-2 ribosomal frameshifting. Despite previous expectations of structural heterogeneity and propensity for multimerization39,40 and despite having a size slightly under that of the previous smallest macromolecule imaged by cryo-EM (28 versus 30 kDa19), the SARS-CoV-2 FSE gave a 6.9-Å resolution monomer map with recognizably helix-like elements arranged into a λ-like tertiary arrangement. Automated secondary structure determination and 3D coordinate building through the recently developed Ribosolve pipeline led to a model with an intricate fold in which stem 1, stem 2 and stem 3 form a closed ring. Notably, the linker connecting the 5′ end of the FSE to stem 1 is threaded through this ring (Fig. 1f). This overall 3D arrangement was tested through a second cryo-EM analysis of an FSE molecule extended by a rationally designed clothespin-shaped nanostructure at stem 3 (Fig. 2). Finally, we have shown the ability of the structure-inspired LNA ASOs to reduce frameshifting efficiencies in in vitro assays (Fig. 3b,e) and to reduce viral replication in cells at 100 nM concentrations (Fig. 3c,d,f). Insights derived from features observed in the cryo-EM structure were important in selecting more appropriate ASO targets and achieving this potency.
Our data and recent results from several groups are revealing a complex picture for SARS-CoV-2 programmed ribosomal frameshifting16,17,40,41. There is accumulating evidence that, in the full SARS-CoV-2 genome context, the FSE can form alternate secondary structures involving genomic segments upstream of the element (Supplementary Fig. 5). As the human ribosome approaches the FSE, it must unfold these upstream structures through its helicase activity, causing the FSE to refold. The position of the 5′ end of the FSE will be influenced by structures that existed prior to ribosome-induced refolding. Depending on this position and the order of formation of stems 1, 2 and 3, the 5′ end could end up threaded through the final FSE stem 1–stem 2–stem 3 ring, as captured by our cryo-EM analysis (Fig. 4). The ring-threaded 5′ end (Fig. 4a) appears poised to lead to ribosomal pausing and −1 PRF through a torsional restraint mechanism11. Indeed, a cryo-EM structure of a stalled mammalian 80S ribosome on an FSE-containing messenger RNA (mRNA) segment has been reported42, and the region identified as the FSE in the density has a near-identical tertiary structure to the one reported here, including the 5′-end threaded topology, supporting the functional relevance of this tertiary fold.
a,b, After the ribosome unfolds upstream genomic structures, the FSE refolds, resulting in a 3D fold in which the 5′ end is either threaded through a ring in the FSE pseudoknot (inset, hole indicated in yellow), as captured in our cryo-EM structure (a) or left unthreaded (b). The model in b reflects an alternate fold with the same pseudoknotted secondary structure but distinct tertiary arrangement, as captured by de novo modeling. The torsional restraint model suggests that the cryo-EM-resolved conformation in a promotes pausing at the slippery site and ribosomal frameshifting and should be targeted for destabilization relative to alternate folds (b) in antiviral efforts seeking to disrupt frameshifting. c, The phyloP score from 119 diverse coronaviruses superimposed over a 3D model of the SARS-CoV-2 FSE, where coloring is centered over the average phyloP score of the entire viral genome (other comparisons are provided in Supplementary Fig. 6).
The structural complexity of FSE 3D folding has implications for therapeutic targeting efforts. Our structural and ASO-targeting results both highlight the importance of the ring-threaded 5′ end of stem 1 for modulating frameshifting. Our top performing ASO, S1D-2, targets this region and shows virus replication inhibition at 100 nM in cell culture, a concentration level that supports testing in animal models43,44,45. Nevertheless, further optimization of S1D-2 is warranted to improve inhibition to low- or even sub-nanomolar concentrations, as ASO length, melting temperature, positions of DNA versus LNA bases, overall ASO nucleotide chemistry and hybridization locations in stem 1 can all affect efficacy and contribute to activity discrepancies between ASOs targeted against even the same region. In addition to structure–activity relationship efforts to identify and characterize lead candidates, evidence of SARS-CoV-2 viral tropism across a range of host cell types has also been shown to directly affect the antiviral response of inhibitors46. These cell type-specific antiviral results are reflective of expression level differences of important host proteins and factors, which are involved in various stages of the viral life-cycle. Our results with EIDD-1931 in different cell types are in line with these findings. Similarly, ASO transfectability differs across cell lines, as evidenced by the variable activity seen across cell types employed in our assays. Utilizing a combination of these cell systems enables a more complete picture of antiviral therapeutic potential.
As an alternative to ASOs, SARS-CoV-2 replication might be disrupted by modulating frameshifting with small molecules. Three potential binding pockets appear in our cryo-EM structure, which we term the ‘ring site’, the ‘J3/2 site’ and the ‘slippery hairpin site’. All three sites involve nucleotides with high sequence conservation (blue, Fig. 4c and Supplementary Fig. 6).
We have provided 3D structural data of a functionally obligate RNA-only segment of the SARS-CoV-2 genome and have described implications for mechanism and targeting of −1 PRF, an intricate and critical genomic process. Dozens of other segments of the 30-kb RNA genome are highly conserved and have been predicted to have their own tertiary structure folds, many in the size range explored here. Applying cryo-EM and antisense targeting to these segments may yield additional information that sheds light on SARS-CoV-2 RNA biology and, hopefully, accelerates the design of genome-disrupting therapeutic agents.
Methods
Sample preparation for chemical mapping and cryo-electron microscopy
The SARS-CoV-2 FSE sequence was derived from the NCBI GenBank reference sequence NC.045512.247 and corroborated using manual alignment against existing annotated betacoronavirus FSEs. For cryo-EM studies, the sequence was then prepended with the Φ2.5 T7 RNA polymerase promoter48. Primers were designed for polymerase chain reaction (PCR) assembly using the Primerize49 web server, ordered from Integrated DNA Technologies, and assembled into full-length double-stranded DNA (dsDNA) by PCR assembly following the Primerize protocol using Phusion polymerase, ‘High-Fidelity’ buffer (Thermo Scientific) and an annealing temperature of 64 °C. Product homogeneity was assessed by 1× TBE (Ambion)—4% agarose gel electrophoresis as visualized with ethidium bromide. The resulting product was purified using Qiagen QIAquick PCR purification kits, eluted in molecular-biology-grade double-distilled H2O (ddH2O, Invitrogen) and quantified using a NanoDrop spectrophotometer (Thermo Scientific). The resulting dsDNA was then used for in vitro transcription (IVT) using a Thermo Scientific TranscriptAid T7 kit (Thermo Scientific) where 22 reactions were carried out at 1.19 μg of dsDNA per reaction and left at 37 °C for 12 h. The resulting product was split equally over four Zymo25 RNA Clean and Concentrator (Zymo Research) columns and cleaned following the manufacturer’s protocol with an extra 15-min drying step before elution in 30 μl of ddH2O per column; RNA was quantified at fivefold dilution via NanoDrop. Next, the RNA was purified by denaturing poly-acrylamide gel electrophoresis (dPAGE), using 15% 29:1 bis-acrylamide, 7 M urea, 1× TBE gels cast in BioRad Criterion 1-mm Midi Cassettes that were pre-run at 25 W for 1 h, before loading the 90 °C denatured RNA in 70% formamide, 1× TBE loading buffer with bromophenol blue and xylene cyanol running markers and run at 25 W for another 45 min. A 50-μl volume of RNA was loaded per gel, so two gels were used. The side wells of each gel were used for size standards and 2/35th diluted RNA for purification corroboration. To prevent potentially disrupting the RNA structure with intercalating dyes, the major RNA products on the dPAGE gels were localized and excised by brief UV shadowing. After the bulk RNA was excised, the remaining gel was stained for 20 min in SyBrGold (Invitrogen) and visualized by UV transillumination. The excised gel pieces were crushed and soaked overnight in 300 μl 10 mM Tris and 1 mM EDTA buffer (Ambion) per gel slice on an active vortexer at 4 °C. The resulting gel slurry was then spin-filtered through 0.45-μm filters (Corning Costar); the filtrate was first purified in one Zymo25 RNA Clean and Concentrator column per excised band, eluting in 40 μl of ddH2O, and then through one Zymo5 RNA Clean and Concentrator column per sample, eluting in 25 μl ddH2O, at which point samples were consolidated and concentrated one last time in one Zymo5 RNA Clean and Concentrator column per sample, eluting in 14 μl ddH2O. RNA was quantified by NanoDrop. The same overall protocol was carried out for the FSE and FSE-ATP-TTR3 chemical mapping constructs (one-dimensional chemical mapping and two-dimensional chemical mapping (M2-seq)). Chemical mapping constructs were prepended with the canonical Φ6.5 T7 RNAP promoter as the 5′ reference hairpins used had 5′ terminal GGs; these constructs were also gel-extracted using blue-light transillumination after staining with SyBrGold. The FSE 1D chemical mapping construct′s assembled dsDNA was 2% agarose gel-purified by size using blue-light transillumination using SyBrSafe (Invitrogen) and purified using a Qiagen MinElute Gel Extraction kit prior to IVT.
Cryo-electron microscopy data acquisition
The RNA samples were folded before plunge freezing as previously described23. Briefly, RNA sample in a buffer containing 50 mM Na-HEPES (pH 8.0) was denatured at 90 °C for 3 min and cooled at room temperature (r.t.) for 10 min; MgCl2 was then added to achieve a final concentration of 15 μM RNA and 10 mM MgCl2, and samples were incubated at 50 °C for 20 min, followed by cooling at r.t. for 3 min. Three microliters of the FSE and FSE-ATP-TTR3 samples were applied onto glow-discharged 200-mesh R2/1 Quantifoil copper grids. The grids were blotted for 4 s and rapidly cryocooled in liquid ethane using a Vitrobot Mark IV instrument (Thermo Fisher Scientific) at 4 °C and 100% humidity. The final datasets in this study were imaged in a Titan Krios cryo-electron microscope (Thermo Fisher Scientific) operated at 300 kV with a GIF energy filter (Gatan) at a magnification of ×165,000 (corresponding to a calibrated sampling of 0.82 Å per pixel) for both samples. Micrographs were recorded by EPU software (Thermo Fisher Scientific) with a Gatan K2 Summit direct electron detector, where each image was composed of 30 individual frames with an exposure time of 6 s and an exposure rate of 8.3 electrons per second per Å2. A total of 10,222 video stacks for FSE and 12,380 video stacks for FSE-ATP-TTR3 were collected.
Single-particle image processing and 3D reconstruction
All micrographs were first imported into Relion for image processing. Motion correction was performed using MotionCor250 and the contrast transfer function (CTF) was determined using CTFFIND451. All particles were autopicked using the NeuralNet option in EMAN224. Then, particle coordinates were imported to Relion25, where the poor 2D class averages were removed by several rounds of 2D classification. The initial models for both datasets were built in cryoSPARC52 using the ab initio reconstruction option. For the FSE, 1,063,711 particles were picked and 445,707 were selected after 2D classification in Relion. After removing classes with poorly connected density by 3D classification, the final 3D refinement was performed using 109,137 particles in Relion, and a 6.9-Å map was obtained. For the FSE-ATP-TTR3, 1,103,091 particles were picked and 902,309 were selected after 2D classification in Relion. After removing classes with poorly connected density by 3D classification in Relion, two rounds of heterogeneous refinement were performed in cryoSPARC to further remove contaminant particles. The final 3D homogeneous refinement was performed using 257,558 particles, and a 6.4-Å map was obtained. Resolution for the final maps was estimated with the 0.143 criterion of the Fourier shell correlation curve without or with mask. A Gaussian low-pass filter was applied to the final 3D maps displayed in the UCSF Chimera software package53.
One-dimensional chemical mapping
Chemical mapping was conducted on FSE, plusFSE and FSE-ATP-TTR3 constructs (Supplementary Table 1), with reference hairpins added to either side of the regions of interest54. In brief, dPAGE-purified RNA was diluted to 80 nM in 50 mM Na-HEPES at pH 8.0 and 10 mM MgCl2, folded using the same folding scheme as described for cryo-EM (plus an extra incubation at 37 °C for 24 min at the end), followed by dilution to 60 nM in either 0.5% dimethyl sulfate (DMS) in 0.5% ethanol in ddH2O or 4.24 mg ml−1 1-methyl-7-nitroisatoic anhydride (1M7) in DMSO, reaction at r.t. for 15 min, followed by quenching in 500 mM NaMES at pH 6.5 for 1M7, and 51% β-mercaptoethanol (β-ME), respectively. During this quench step, fluorescein amidite (FAM)-labeled Tail2 reverse transcription primer was added to a final concentration of 2 nM, with the RNA at 40 nM. Reverse transcription was then carried out using SuperScript III (Invitrogen) alongside an RNA dideoxynucleotide (ddNTP) sequencing ladder. The resulting complementary DNA (cDNA) was cleaned and then resuspended in HiDi-formamide (Applied Biosystems) with ROX350 (Applied Biosystems) added as fiducial markers. Concentrated and dilute samples were then submitted for capillary electrophoresis at Elim Biopharmaceuticals and the resulting FAM channel traces, as aligned by the ROX350 markers, were used in HiTRACE52 to assign per-nucleotide reactivity data for DMS, 1M7 and negative control incubation conditions. The resulting .rdat files were then processed using RNAStructure with 100 bootstrapping rounds, allowing for the computation of pseudoknots with Biers software (https://github.com/ribokit/Biers).
Two-dimensional chemical mapping (M2-seq)
Following 1D chemical mapping, the same constructs were also subjected to 2D chemical mapping, which uses DMS modification on folded RNA but leverages information on structural perturbations from sequence mutations to more directly infer helices55. The resulting signals were read out using Illumina short read sequencing, exploiting the mutational readthrough of DMS modified bases by the retrotranscriptase TGIRT-III (InGex).
In brief, the same DNA encoding the 1D chemical mapping constructs was first subjected to error-prone PCR (epPCR), where 2 ng μl−1 of PCR assembled (see ‘Sample preparation for chemical mapping and cryo-electron microscopy’) dsDNA (1.6 μg total dsDNA used per construct, 8× epPCRs) was amplified using the epPCR forward and reverse primers at 100 μM (Supplementary Table 1) under error-prone conditions (10 mM Tris-HCl pH 8.3, 50 mM KCl, 7 mM MgCl2, 1 mM dTTP, 1 mM dCTP, 200 nM dATP, 200 nM dGTP, 500 nM MnCl2 and 1× Taq polymerase (New England BioLabs)), using the following PCR cycling conditions: initial denaturation at 98 °C for 60 s; then 35 cycles of 94 °C for 30 s, 60 °C for 60 s and 72 °C for 180 s; followed by 72 °C for 10 min. The resulting PCR products were concentrated using Qiagen QIAquick PCR purification kits (one per 200 μl of product). The resulting cleaned dsDNA was purified by size on a SyBrSafe-stained 2% agarose gel using blue-light transillumination, and purified using a Qiagen MinElute Gel Extraction kit before IVT (4× TranscriptAid reactions, 2,000 ng of dsDNA per reaction, 12-h reaction at 37 °C followed by 20 min of DNase I treatment at 37 °C, 1 μl per reaction volume). The resulting RNA was Zymo25-purified and then 8% dPAGE-purified by size using SyBrGold, blue-light transillumination, and one ZR small-RNA PAGE Recovery kit per construct following the manufacturer’s protocol. The resulting size-purified RNA was then subjected to the M2-seq protocol. Briefly, unmodified (nomod) and DMS-modified samples were prepared, using 5 pmol and 12.5 pmol of RNA per condition, respectively. RNA was diluted into 10 mM Tris, 1 mM EDTA buffer (Ambion) to 3.0 μl, denatured at 90 °C for 3 min, cooled to r.t. for 10 min, and then diluted into 5.0 μl of 1.5 M sodium-cacodylate pH 7.0, 50 mM MgCl2 buffer and 14.5 μl ddH2O and allowed to refold at 50 °C for 20 min followed by cooling to r.t. for 3 min. Following this folding, the buffered RNA was then subjected to DMS chemical modification in 2.5 μl of 15% DMS (diluted in 100% ethanol) or just 85% ethanol for the no modification condition, at 37 °C for 6 min, followed by quenching with 25.0 μl of β-mercaptoethanol. All of the DMS and β-mercaptoethanol steps were carried out in a fume hood. The resulting quenched RNA reactions were then diluted in ddH2O to 100 μl and ethanol-precipitated using GlycoBlue (Invitrogen) as a co-precipitant, resuspending the resulting RNA in 7.0 μl of ddH2O. Next, the cleaned nomod and DMS-modified RNA was reverse-transcribed using reverse transcription barcode (RTB) FAM-labeled primers (Supplementary Table 1) using a unique index per condition, per construct. A 4.6 μl volume of RNA was incubated with 0.930 μl of 285 nM RTB primer in 6.520 μl of retrotranscription buffer: 2.4 μl 5× TGIRT-III buffer, 1.2 μl 10 mM dNTPs mix (New England BioLabs), 0.6 μl 100 mM DDT (Invitrogen), 1.32 μl of ddH2O and 1.0 μl of TGIRT-III enzyme. The reactions were allowed to sit at 25 °C for 5 min, before being kept at 64 °C for 3 h. Afterwards, the reactions were treated with 5.0 μl 400 mM NaOH at 90 °C for 3 min, acid-quenched (see above) with 2.2 μl of quench acid, and diluted in 30.75 μl of ddH2O before purification using a Zymo Oligo Clean and Concentrator kit, eluting in 12.5 μl of ddH2O. The resulting cleaned cDNA was then individually subjected to unidirectional PCR for second-strand synthesis using the second-strand synthesis primer (Supplementary Table 1). A 1.0 μl volume of 100 μM primer was used per 2.5 μl of cDNA, and 19.5 μl of PCR master mix (same as PCR assembly, see above). The reaction mixtures were initially denatured at 98 °C for 60 s; then four cycles of 98 °C for 10 s, 58 °C for 30 s and 72 °C for 30 s; followed by 72 °C for 10 min. Immediately after the final step, PCR amplification using the iTru p5 and p7 primers (Supplementary Table 1) was conducted in the same reaction mixtures. A 2.0 μl volume of equimolar p5/p7 (50 μM per primer) mixture was spiked into each unidirectional PCR reaction product, mixed, spun down and quickly returned to the thermal cycler for the following: initial denaturation at 98 °C for 60 s; then 5× cycles of 98 °C for 10 s, 50 °C for 30 s and 72 °C for 30 s; followed by 15 cycles of 98 °C for 10 s, 60 °C for 30 s, increasing this temperature incrementally each cycle to a final temperature of 70 °C by cycle 15, and 72 °C for 30 s; then 72 °C for 10 min. The resulting reaction products were concentrated using Qiagen QIAquick PCR purification kits, and finally 1% agarose gel-purified by size using blue-light transillumination using SyBrSafe and purified using Qiagen MinElute Gel Extraction kits. The resulting dsDNA sequencing libraries were then sequenced using an Illumina MiSeq system using the v3 600 cycle kit, loading equimolar amounts of DMS-treated DNA per construct, and threefold less nomod DNA per construct (14 fmol of dsDNA total including 40% PhiX control).
M2-seq data analysis was performed using the pipeline at https://github.com/ribokit/M2seq. FASTQ files were demultiplexed with NovoBarcode, and demultiplexed files were processed with ShapeMapper to obtain mutation strings for each read in a binary format. The simple_to_rdat.py script was used to generate 2D mutational profiles in RDAT files. These files were then analyzed with Biers (https://ribokit.github.io/Biers/) to generate Z-scores, normalized 1D DMS profiles and secondary-structure predictions using ShapeKnots with 100 bootstrapping iterations guided by both Z-scores and 1D DMS profiles.
Reactivity-guided structure modeling for extended FSE
Secondary structure modeling was performed for FSE constructs including 110 upstream nucleotides, guided by SHAPE reactivity data collected in this study, along with various recently published SHAPE and DMS reactivity datasets56. SHAPE reactivity data were collected for the extended FSE constructs with 1M7 as described in ‘Two-dimensional chemical mapping (M2-seq)’, and data were averaged across two replicates. DMS reactivity data were normalized by scaling the median of the top 5% of reactive bases to 1.0 and winsorizing. The predictions reported here were made with ShapeKnots and 100 bootstrapping iterations if pseudoknots were predicted; otherwise, RNAStructure’s Fold algorithm was used, again with 100 bootstrapping iterations. All secondary structures were depicted using RiboDraw (https://github.com/ribokit/RiboDraw/).
FSE model building
The 6.9-Å cryo-EM FSE map was first subjected to segmentation using Segger31 to remove any contours lower than 0. The resulting segmented map was then low-pass-filtered to 20-Å by auto-DRRAFTER, and a map threshold of 0.05 was used to compute the auto-DRRAFTER fitting nodes. Auto-DRRAFTER identifies graph end nodes in both the experimental cryo-EM map and the secondary structure, and then seeks to match these end nodes together to initiate model building. Because the FSE is expected to have only one stem loop (one end node), the SARS-CoV-2 FSE stem 311, stem 3 was hence used by the software as the initial point for docking the secondary structure into the cryo-EM map. Auto-DRRAFTER identified two map end nodes (Extended Data Fig. 3f) into which this stem 3 was automatically docked, and both were used for two identical and converged runs using stem 3 as the docked helix, using the literature secondary structure as a constraint (Supplementary Table 1). We noted that jobs explicitly initiated with stem 3 docking into what was later determined to be the incorrect region of the density (using the FSE-ATP-TTR-3) produced results very similar to jobs initiated with the other, correct, node used as the docking point (Extended Data Fig. 3g). After this set of modelling as well as the cross-validation using FSE-ATP-TTR-3, subsequent auto-DRRAFTER jobs were conducted only using the correct node as the specified docking position for stem 3. Ultimately, jobs run using the 1D chemical mapping-derived secondary structure resulted in marginally (~0.8 Å lower estimated accuracy) better correlated models compared to models generated with either the literature or M2-seq secondary structures (Extended Data Fig. 3) and so were used for subsequent analysis. Regardless of the starting secondary structure, all auto-DRRAFTER runs resulted in similar global tertiary structures (Extended Data Fig. 3). Modelling was attempted using the FSE-ATP-TTR3 map, using the previously computed ATP-TTR 3 model as the initial docking structure, but the absence of clear density at the FSE portion of the map produced convergent results that only confidently placed the stem 3 helix (Fig. 2c). All jobs were run on the Stanford high-performance computing cluster, Sherlock 2.0, using the latest distribution of Rosetta and auto-DRRAFTER.
FSE-ATP-TTR3 design
ATP-TTR-3, a stabilized aptamer for ATP and AMP, was selected as a tag due to its stability, structural features conducive to projection angle identification and its helical end for linkage. The tag was linked to the FSE by removing the dimerization loop (44–52) and placing the tag in its place. Nucleotides were inserted in the FSE-tag junction (C44G and U52A, FSE numbering) to remove any predicted secondary-structure interactions between the FSE and tag and retain the individual secondary structure prediction for FSE and the ATP-TTR 3 tag. Secondary structure was predicted using CONTRAfold 2.057 using the command ‘contrafold predict seq.fasta’.
FSE-ATP-TTR3 density fitting
The previously obtained density of ATP-TTR 3 and the FSE density were fit into the density obtained for the FSE-ATP-TTR3 construct using Situs32. The overlap was not expected to be ideal, because the FSE-ATP-TTR3 construct does not contain the dimerization loop of the FSE (nucleotides 44–52) and has extra bases, C44 and U175, inserted at the fusion interface between the FSE and ATP-TTR 3, a linkage not present in either individual construct. First, the ATP-TTR 3 density was fit into the FSE-ATP-TTR3 density using the following commands:
vol2pdb ATP-TTR_apo_ribosolve.mrc atp.pdb
colores FSE-ATP-TTR3.mrc atp.pdb -res 4
Next, the docking result with the highest correlation was subtracted from the FSE-ATP-TTR3 density:
pdb2vol atp_best_001.pdb atp_best_001.mrc #using 1A voxel spacing
voldiff FSE-ATP-TTR3.mrc atp_best_001.mrc diff.mrc
Finally, the FSE density was fit into the difference density:
vol2pdb FSE-wt.mrc fse.pdb
colores diff.mrc fse.pdb -res 7
pdb2vol col_best_001.pdb fse_001.mrc #same for all 9 hits, using 1A voxel spacing
The results were visualized in Chimera. The stem attached to the ATP-TTR 3 on the docking result with the highest correlation was assigned to stem 3.
De novo RNA 3D structure modeling
De novo models were generated for versions of the SARS-CoV-2 frameshift stimulation element with 2, 6, 10 or 14 bp in stem 1, along with frameshift stimulation elements from seven betacoronaviruses. Models were obtained using Rosetta′s rna_denovo application using the standard FARFAR2 protocol58. For stem 1 variants of the SARS-CoV-2 frameshift stimulation element, shortened stems were truncated by removing base pairs from the 5′ end of the 5′ strand of stem 1, and longer stems were formed by adding additional GC base pairs to the 5′ end of the 5′ strand of stem 1. Secondary structures for the frameshift stimulation elements for MHV, human coronavirus HKU1, human coronavirus OC43, BCoV, MERS and SARS-CoV-1 were obtained from previous literature analyses11. For MERS, 2,000 decoys were produced using the FARFAR2 ROSIE server. De novo models for the SARS-CoV-2 FSE models were obtained as described previously59. For SARS-CoV-2 frameshift stimulation elements varying stem 1, 20,000–30,000 decoys were generated using the Stanford high-performance computing cluster, Sherlock 2.0; for all other cases, 150,000 decoys were generated with the Open Science Grid. In each case, the 400 top-scoring models were clustered using the rna_cluster Rosetta application with a 5-Å cluster radius. From the top 10 clusters, top-scoring cluster members demonstrating the threaded and unthreaded topologies were selected. Models are available at https://github.com/DasLab/FARFAR2_FSE_homology.
phyloP analysis
phyloP scores were extracted directly from the UCSC SARS-CoV-2 genome browser for both the whole genome and the FSE.
LNA design and preparation
LNAs are oligonucleotides with a 2′ sugar modification that locks the ribose in a C3′-endo conformation by a 2′-O, 4′-C methylene bridge, which in turn locks the LNA into the A-form helical conformation60. Oligonucleotides containing LNAs were custom-synthesized by Integrated DNA Technologies, purified by HPLC, and resuspended in ddH2O. (+) denotes LNA base. All others represent (non-locked) DNA nucleotides. Select LNA ASOs were made with phosphorothioate (PS) internucleotide linkages as indicated in the following. LNA ASOs were designed to be 14–16 nucleotides in length, with the LNA gapmer ASOs containing three or four LNA bases at the 5′ and 3′ termini, flanking the internal DNA sequence. The accessibility of the target RNA structure to antisense oligonucleotide binding, target sequence GC content, ASO-RNA hybridization scoring, and melting temperature (Tm) were taken into account for the LNA design (https://www.qiagen.com/us/service-and-support/learning-hub/technologies-and-research-topics/lna/custom-lna-design-and-applications/custom-lna-oligo-design-guidelines/).
S2D (stem 2 disruptor): 5′ +CCCTG+TA+TA+CGACA+T 3′
S3D-1 (stem 3 disruptor): 5′ +A+C+ATCAGTACT+A+G+T 3′, PS linkages
S3D-2 (stem 3 disruptor-2): 5′ +T+C+AGTACTAGTG+C+C+T+G 3′, PS linkages
Slp2 (slippery site 2): 5′ +A+C+C+GCGAACCCGTT+T+A+A 3′, PS linkages
S1D-1 (stem 1 disruptor-1): 5′ +C+G+CACGGTGTA+A+G+A+C 3′, PS linkages
S1D-2 (stem 1 disruptor-2): 5′ +T+A+AGACGGGCTGC+A+C+T 3′, PS linkages
S1D-3 (stem 1 disruptor-3): 5′ +C+A+C+TTACACCGCA+A+A+C 3′, PS linkages
Negative control LNA ASOs:
Scramble LNA (Scr. LNA): 5′ C+ATAC+GTC+TAT+AC+GCT 3′, PS linkages
JL4: 5′ +CCCTG+TAAA+ACGGG+C 3′
RL4: 5′ +CGGGC+TAAA+AGCCC+T 3′
ASO control for caspase assay:
Pos_LNA: 5′ +A+G+C+CAGACAG+C+G+A 3′, PS linkages
The JL4 and RL4 ASOs contained short regions of hybridization to stem 1 and stem 2, but were not expected to be able to displace intramolecular pairings in those stems.
In vitro frameshifting assays
Frameshifting levels for SARS-CoV-2 were determined using the p2luc bicistronic dual-luciferase reporter assay system. The SARS-CoV-2 frameshift stimulation element sequence (GUUUUUAAACGGGUUUGCGGUGUAAGUGCAGCCCGUCUUACACCGUGCGGCACAGGCACUAGUACUGAUGUCGUAUACAGGGCUUUUGAU) was inserted into the p2luc vector via the BamHI and SacI restriction sites, such that it was located between the Renilla (Rluc) and Firefly (Fluc) luciferases, with Fluc located in the −1 frame relative to Rluc. A control construct was also constructed with a single-nucleotide insertion in addition to a disrupted slippery sequence, which causes Fluc to be in the 0 frame relative to Rluc and allows for the normalization of frameshifting readings to 100% (GUUAUUCAAGCGGGUUUGCGGUGUAAGUGCAGCCCGUCUUACACCGUGCGGCACAGGCACUAGUACUGAUGUCGUAUACAGGGCUUUUGAU). The HIV frameshifting element was used as a control (slippery, UUUUUUAGGGAAGAUCUGGCCUUCCCACAAGGGAAGGCCAGGGA; control, CUUCUUAAGGGAAGAUCUGGCCUUCCCACAAGGGAAGGCCAGGGA).
mRNA was transcribed using the mMESSAGE mMACHINE T7 transcription kit (Invitrogen) and purified using the MEGAclear Transcription Clean-Up kit, following the manufacturers’ protocols. Triplicates of 200 ng of both mRNAs were mixed with 2.5 µl of 80, 40, 20, 8, 4, 2 and 0 µM LNA for 1 h at r.t. In vitro translation assays were run on the prepared mRNA templates using Rabbit Reticulocyte Lysate kits (Promega) at a final volume of 25 µl, by incubating at 30 °C for 1.5 h. Luciferase readings were performed using the Dual Luciferase Reporter Assay system (Promega) on a Biotek Synergy Neo2 plate reader, then 3 × 2 μl volumes of lysate in three separate wells were measured for each prepared translation replicate (Supplementary Table 2).
Frameshifting levels were calculated by the ratio (in which lucS refers to the SARS-CoV-2 FSE sequence, and lucC to the disrupted slippery sequence control):
The resulting mean fold-change frameshifting data were then fitted, using the MATLAB 2019b (MathWorks) ‘Trust Region’ algorithm, to a standard binding isotherm with the estimated half maximal inhibitory concentration (IC50) constrained to all numbers between 0 and 8 µM:
LNA treatment, caspase activation and cell viability assays
All cell lines used for in cellulo experiments, Vero E6 and Huh-7 and ACE2-A549, were maintained at 37 °C in complete DMEM (Gibco) containing 10% FBS (Invitrogen), penicillin and streptomycin (Gibco), and HEPES buffer (Gibco). The ACE2-A549 cells were specially engineered to over-express the ACE2 receptor in human alveolar basal epithelial cells (A549).
For caspase activation and cell viability assays, one day before transfection, 10,000 cells per well were plated in growth medium in black-walled, clear-bottom 96-well plates (EK-25090, E&K Scientific) so as to be 70% confluent at the time of transfection. After 24 h, LNA-modified ASOs were diluted in Opti-MEM I and mixed with Lipofectamine 3000 reagent (Life Technologies) according to the manufacturer’s protocol to the indicated final concentrations. The complexes were incubated at r.t. for 30 min, after which 10 μl of each LNA–Lipofectamine complex was added dropwise to cells containing 90 μl of growth medium. Cells were then incubated as described. A caspase-activating LNA, Pos_LNA, and known apoptotic activator, camptothecin, were used as positive controls.
To test caspase activation, 24 h post-transfection with the LNA ASOs, Caspase-Glo 3/7 reagent (G8092/G8093 kit; Promega) was prepared at r.t. according to the manufacturer’s specifications. Once the cell plates were equilibrated at r.t., a 1:1 ratio of Caspase-Glo 3/7 reagent volume to sample volume was added, then its contents were mixed gently at 350–500 r.p.m. for 30 s using a plate shaker, and left to incubate at r.t. for 45 min in the dark. The plates were measured for caspase activation by luminescence using a Tecan Infinite multimode plate reader (M1000, Tecan). Data were analyzed and graphed in Prism 8 by GraphPad.
Cell viability after LNA ASO treatment was determined by PrestoBlue cell viability reagent (Invitrogen) following the manufacturer’s instructions. LNA ASO transfections were generated using the same transfection protocol as described above in 96-well plates. At 24 h post-transfection, the medium was replaced with complete DMEM and cells were incubated for an additional three days, then 10 μl of Prestoblue reagent (cat. no. 11644807; Roche Applied Science) was added to each well and the plate incubated for 1 h. The plate was then read for fluorescence (540-nm excitation/590-nm emission) using a Tecan Infinite multimode plate reader (M1000). Cell viability after four days of LNA ASO treatment was calculated as a percentage of total cellular viability, normalized to the average viability count of non-treated wells in the absence of LNA ASOs after background correction. Data were analyzed and graphed in Prism 8 by GraphPad.
Cellular SARS-CoV-2 replication assays
For cellular replication assays, LNA ASOs were reconstituted in RNase-free water to 100 μM stock solutions, aliquoted and stored at −20 °C before single use. One day before transfection, Huh-7, Vero E6 or ACE-A549 cells were plated in 96-well clear-bottom plates to 60–70% confluency at the time of treatment with the LNA ASOs S2D, S3D-1, S3D-2 or scrambled LNA. Lipofectamine 3000 (Life Technologies) was used to transfect LNA ASOs into cells at 25 nM or 100 nM final concentration, according to the manufacturer’s protocol. Cells were then infected with SARS-CoV-2 reporter virus expressing nanoluciferase (SARS-CoV-2 nLUC) at a multiplicity of infection of 0.3 for 1 h, after which the virus was removed and fresh medium was added. Recombinant SARS-CoV-2 nLUC is an authentic fully replicating virus in which ORF7 has been deleted and replaced with nLUC. Thus, the measurement of nLUC expression is a surrogate marker of virus replication enabling the screening of antiviral compounds, obviating the need for costly and time-consuming virus titering assays. A nucleoside analog NHC (a.k.a. EIDD-1931) with potent activity against SARS-CoV-2 was included as a positive control. A DMSO control was included as a mock-treated, negative control. Data were graphed and analyzed in Prism 8 by GraphPad. Statistical analysis of the data from each cell type was computed as an ordinary one-way analysis of variance using Dunnet’s multiple comparisons test against the DMSO control or scrambled LNA control from each cell type, where indicated.
Correlation analysis between in vitro and in cellulo assays
To calculate the correlation between the dual luciferase in vitro assays and the in cellulo viral replication assays, −1 PRF efficiencies were computed using the binding isotherm fits (see in vitro frameshifting methods above) at the LNA concentrations used for the cell culture experiments, 25 and 100 nM (Extended Data Fig. 9). These −1 PRF efficiencies were then compared against the scrambled LNA normalized change in observed log10(luciferase) values using the linear Spearman correlation calculation in GraphPad Prism 8. Over the whole dataset, the strongest correlation appeared to arise from the concentration, and not the identity, of the LNAs used, with r2 values for the aggregate data of 0.45 (P > 0.0025), 0.00 for the 25 nM samples (P > 0.9369) and 0.14 for the 100 nM samples (P > 0.4709).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this Article.
Data availability
Cryo-EM maps of the FSE and FSE-ATP-TTR3 have been deposited in the Electron Microscopy Data Bank under accession codes EMD-22296 and EMD-22297; atomic models of FSE have been deposited in the Protein Data Bank under accession code 6XRZ. Source data are provided with this paper.
Code availability
HiTRACE and M2seq codes are available from: https://ribokit.github.io/HiTRACE/ and https://ribokit.github.io/M2seq/.
References
Mullard, A. Flooded by the torrent: the COVID-19 drug pipeline. Lancet 395, 1245–1246 (2020).
Batool, M., Ahmad, B. & Choi, S. A structure-based drug discovery paradigm. Int. J. Mol. Sci. 20, 2783 (2019).
Cully, M. Antifungal drugs: small molecules targeting a tertiary RNA structure fight fungi. Nat. Rev. Drug Discov. 17, 864 (2018).
Burnett, J. C. & Rossi, J. J. RNA-based therapeutics: current progress and future prospects. Chem. Biol. 19, 60–71 (2012).
Walls, A. C. et al. Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292 (2020).
Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020).
de Wit, E., van Doremalen, N., Falzarano, D. & Munster, V. J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 14, 523–534 (2016).
Plant, E. P., Rakauskaite, R., Taylor, D. R. & Dinman, J. D. Achieving a golden mean: mechanisms by which coronaviruses ensure synthesis of the correct stoichiometric ratios of viral proteins. J. Virol. 84, 4330–4340 (2010).
Dinman, J. D. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip. Rev. RNA 3, 661–673 (2012).
Marra, M. A. The genome sequence of the SARS-associated coronavirus. Science 300, 1399–1404 (2003).
Plant, E. P. et al. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol. 3, e172 (2005).
Dieckmann, A. et al. A sensitive in vitro approach to assess the hybridization-dependent toxic potential of high affinity gapmer oligonucleotides. Mol. Ther. Nucleic Acids 10, 45–54 (2018).
Plant, E. P., Sims, A. C., Baric, R. S., Dinman, J. D. & Taylor, D. R. Altering SARS coronavirus frameshift efficiency affects genomic and subgenomic RNA production. Viruses 5, 279–294 (2013).
Wacker, A. et al. Secondary structure determination of conserved SARS-CoV-2 RNA elements by NMR spectroscopy. Nucleic Acids Res. 48, 12415–12435 (2020).
Hansen, T. M., Reihani, S. N. S., Oddershede, L. B. & Sørensen, M. A. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc. Natl Acad. Sci. USA 104, 5830–5835 (2007).
Ahn, D.-G. et al. Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication. Antivir. Res. 91, 1–10 (2011).
Park, S.-J., Kim, Y.-G. & Park, H.-J. Identification of RNA pseudoknot-binding ligand that inhibits the −1 ribosomal frameshifting of SARS-coronavirus by structure-based virtual screening. J. Am. Chem. Soc. 133, 10094–10100 (2011).
Haniff, H. S. et al. Targeting the SARS-CoV-2 RNA genome with small molecule binders and ribonuclease targeting chimera (RIBOTAC) degraders. ACS Cent. Sci. 6, 1713–1721 (2020).
Zhang, K. et al. Structure of the 30-kDa HIV-1 RNA dimerization signal by a hybrid cryo-EM, NMR and molecular dynamics approach. Structure 26, 490–498 (2018).
Guo, S. et al. Ultra-thermostable RNA nanoparticles for solubilizing and high-yield loading of paclitaxel for breast cancer therapy. Nat. Commun. 11, 972 (2020).
Zhang, K. et al. Cryo-EM structure of a 40-kDa SAM-IV riboswitch RNA at 3.7 Å resolution. Nat. Commun. 10, 5511 (2019).
Li, S. et al. Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions. Nat. Struct. Mol. Biol. 26, 1094–1105 (2019).
Kappel, K. et al. Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures. Nat. Methods 17, 699–707 (2020).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Liao, H. Y. & Frank, J. Definition and estimation of resolution in single-particle reconstructions. Structure 18, 768–775 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Spahn, C. M. T., Grassucci, R. A., Penczek, P. & Frank, J. Direct three-dimensional localization and positive identification of RNA helices within the ribosome by means of genetic tagging and cyro-electron microscopy. Structure 7, 1567–1573 (1999).
Yesselman, J. D. et al. Computational design of three-dimensional RNA structure and function. Nat. Nanotechnol. 14, 866–873 (2019).
Pintilie, G. D., Zhang, J., Goddard, T. D., Chiu, W. & Gossard, D. C. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170, 427–438 (2010).
Kovacs, J. A., Galkin, V. E. & Wriggers, W. Accurate flexible refinement of atomic models against medium-resolution cryo-EM maps using damped dynamics. BMC Struct. Biol. 18, 12 (2018).
Papargyri, N., Pontoppidan, M., Andersen, M. R., Koch, T. & Hagedorn, P. H. Chemical diversity of locked nucleic acid-modified antisense oligonucleotides allows optimization of pharmaceutical properties. Mol. Ther. Nucleic Acids 19, 706–717 (2020).
Lima, W. F., Vickers, T. A., Nichols, J., Li, C. & Crooke, S. T. Defining the factors that contribute to on-target specificity of antisense oligonucleotides. PLoS ONE 9, e101752 (2014).
Burdick, A. D. et al. Sequence motifs associated with hepatotoxicity of locked nucleic acid—modified antisense oligonucleotides. Nucleic Acids Res. 42, 4882–4891 (2014).
Sheahan, T. P. et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 12, eabb5883 (2020).
Plant, E. P. & Dinman, J. D. Torsional restraint: a new twist on frameshifting pseudoknots. Nucleic Acids Res. 33, 1825–1833 (2005).
Hou, Y. J. et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468 (2020).
Ishimaru, D. et al. RNA dimerization plays a role in ribosomal frameshifting of the SARS coronavirus. Nucleic Acids Res. 41, 2594–2608 (2013).
Kelly, J. A. et al. Structural and functional conservation of the programmed −1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2). J. Biol. Chem. 295, 10741–10748 (2020).
Neuman, B. W. et al. Inhibition, escape and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J. Virol. 79, 9665–9676 (2005).
Bhatt, P. R. et al. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science 372, 1306–1313 (2021).
Di Martino, M. T. et al. In vitro and in vivo activity of a novel locked nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma cells. PLoS ONE 9, e89659 (2014).
Jiang, X. et al. Targeting hepatic miR-221/222 for therapeutic intervention of nonalcoholic steatohepatitis in mice. EBioMedicine 37, 307–321 (2018).
Correction for Lim et al., Inhibition of DUX4 expression with antisense LNA gapmers as a therapy for facioscapulohumeral muscular dystrophy. Proc. Natl. Acad. Sci. USA 117, 21823 (2020).
Shin, D. et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 587, 657–662 (2020).
Wu, F. et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020).
Coleman, T. M., Wang, G. & Huang, F. Superior 5′ homogeneity of RNA from ATP-initiated transcription under the T7 φ2.5 promoter. Nucleic Acids Res. 32, e14 (2004).
Tian, S., Yesselman, J. D., Cordero, P. & Das, R. Primerize: automated primer assembly for transcribing non-coding RNA domains. Nucleic Acids Res. 43, W522–W526 (2015).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Yoon, S. et al. HiTRACE: high-throughput robust analysis for capillary electrophoresis. Bioinformatics 27, 1798–1805 (2011).
Cheng, C. Y., Kladwang, W., Yesselman, J. D. & Das, R. RNA structure inference through chemical mapping after accidental or intentional mutations. Proc. Natl Acad. Sci. USA 114, 9876–9881 (2017).
Manfredonia, I. et al. Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Res. 48, 12436–12452 (2020).
Do, C. B., Woods, D. A. & Batzoglou, S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics 22, e90–e98 (2006).
Watkins, A. M., Rangan, R. & Das, R. FARFAR2: improved de novo Rosetta prediction of complex global RNA folds. Structure 28, 963–976 (2020).
Rangan, R., Watkins, A. M., Kladwang, W. & Das, R. De novo 3D models of SARS-CoV-2 RNA elements from consensus experimental secondary structures. Nucleic Acids Res. 49, 3092–3108 (2021).
Hagedorn, P. H. et al. Locked nucleic acid: modality, diversity and drug discovery. Drug Discov. Today 23, 101–114 (2018).
Acknowledgements
We thank the administrative staff of the Biochemistry Department at Stanford, the SLAC National Accelerator Laboratory and the Stanford University COVID-19 research oversight committee for supporting conduct of these studies during a university-wide shutdown. We thank P. Berg for extensive discussion and advice, C. Hecksel and P. Mitchell for expert maintenance of Stanford-SLAC Cryo-EM facilities, K. Kappel for advice on using auto-DRRAFTER, H. Berman and B. Vallat for RNA model validation, M. Elazar for advice on viral replication and cytotoxicity assays, V. Topkar for advice and assistance with M2-seq, A. Watkins for running de novo modeling on the Open Science Grid, A. Siepel and R. Ramani for assistance in interpreting the phyloP data, J. Puglisi for draft review, discussion and advice, J. Kong and C. Patel for providing reagents for PAGE gels and Ban and Atkins laboratories for sharing their preliminary cryo-EM data of their stalled 80S ribosome on a SARS-CoV-2 FSE containing mRNA. This work was supported by National Institutes of Health grants (S10OD021600, R01AI148382, P01AI120943, R01GM079429, P41GM103832, U24GM129564 to W.C.; R35GM122579 to R.D; 1R01AI13219101 to J.S.G.), the DOE Office of Science through the National Virtual Biotechnology Laboratory, a consortium of DOE national laboratories focused on the response to COVID-19, with funding provided by the Coronavirus CARES Act (W.C. and R.D.); the National Science Foundation Graduate Research Fellowship Program grant no. 1650114 (R.R.); NIH/NIDDK 5T32DK007056-44 (E.A.P.); a Stanford ChEM-H Physician Scientist Research Fellowship (E.A.P.); a Stanford ChEM-H COVID-19 Drug and Vaccine Prototyping seed grant (P. Berg, R.D., J.S.G., W.C.); a Stanford ChEM-H Postdocs at the Interface seed grant (R.J.H.); USAMRAA DOD W81XWH1810647 (J.S.G.), Harrington Scholar Innovator Grant (J.S.G.) and Open Philanthropy (J.S.G.); and a Stanford Paul and Mildred Berg Graduate Fellowship (I.N.Z.).
Author information
Authors and Affiliations
Contributions
W.C., R.D., J.S.G., K.Z., I.N.Z. and R.J.H. conceived the study and designed the experiments. I.N.Z. prepared the RNA samples, performed the chemical mapping and built the models. K.Z. performed cryo-EM sample preparation, screening, data collection, image processing and structure determination. S.L. assisted with cryo-EM image processing. R.K. designed the FSE-ATP-TTR3 RNA construct and performed the density fitting. I.N.Z. designed S2D, RL4 and JL4. R.J.H. designed all other LNAs, with input from E.A.P. Y.J.H. made and titered the recombinant SARS-CoV-2 nLUC virus used in the studies. R.J.H. and T.P.S. performed in cellulo assays. M.W. and R.H. performed in vitro frameshifting assays. G.D.P. and R.K. made videos and carried out cryo-EM correlation analysis. R.R. ran RNAStructure analysis, compiled published data from external laboratories for assessment, and carried out de novo modeling. W.K. conducted the FSE-LNA binding assay with replicates and validation by C.B.-S. W.C., R.D., J.S.G., K.Z., I.N.Z., R.J.H., R.R., S.L., G.D.P., M.W., R.H. and V.D. analyzed the data. K.Z., I.N.Z., R.J.H., W.C. and R.D. wrote and edited the manuscript with input from all other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Structural & Molecular Biology thanks Jonathan Dinman, Jane Richardson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Florian Ullrich was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Single-particle cryo-EM analysis of the FSE.
a. Workflow of cryo-EM data processing of FSE. b. Resolution map for the final 3D reconstruction.
Extended Data Fig. 2 Secondary structure of the SARS-CoV-2 Frameshift Stimulation Element (FSE).
a. M2-seq Z-score plot for the 88 nt FSE, with numbering convention as used in the main text. b. Bootstrap confidence values for each base-pair, obtained from ShapeKnots guided by M2-seq Z-scores and the 1D DMS chemical mapping signal from M2-seq. Key structural features are indicated. c. Secondary structure of the FSE, as determined by using ShapeKnots guided by M2-seq Z-scores and the 1D DMS chemical mapping signal from M2-seq. Bootstrapping (100 iterations) support for each continuous helix is shown as an underlined percentage. Nucleotides are colored by 1D DMS chemical mapping signal, and nucleotides unreactive to DMS are depicted in grey. Inset depicts literature secondary structure. d. Secondary structure of the FSE as determined by 1D chemical SHAPE mapping with SHAPE reactivity data overlaid. Structure shown here is computed from ShapeKnots allowing for pseudoknots. Bootstrapping (100 iterations) support for each continuous helix is shown as an underlined percentage. e. Secondary structure as determined by 1D SHAPE mapping with DMS reactivity data overlaid. Helices that are also predicted with ShapeKnots guided by DMS mapping have bootstrapping support indicated.
Extended Data Fig. 3 Consistent ‘threaded’ tertiary structures of the SARS-CoV-2 Frameshift Stimulation Element (FSE) as guided by the 6.9-Å resolution map using autoDRRAFTER.
Over five different starting secondary structures with the specific used secondary structure (left), ensemble of the final, converged, autoDRRAFTER results with final mean pairwise root mean squared distance (RMSD) equivalent to an estimated accuracy in Å indicated (middle), and a simplified view indicating the threading event seen in all results (right). Yellow circles indicate the rings observed in each modelling run. a. the ‘literature’ secondary structure. b. the secondary structure as determined by 1D chemical mapping using 1M7 (see Methods). c. the secondary structure as determined by 2D chemical mapping using DMS (‘M2-seq’, see Methods). d. the M2-seq secondary structure truncated partially through the Slippery Site to better match the observed cryo-EM density. e. the M2-seq secondary structure truncated at both terminal unstructured ends. f. the map indicating the two end-nodes identified by autoDRRAFTER, the correct Stem 3 node (yellow), and the incorrect node for Stem 3 (red). g - left. an early round of autoDRRAFTER modelling after forcing an initial placement of Stem 3 (using the ‘literature’ secondary structure, see below) at the wrong (red) node, note how all results are poorly fitted inside of the map. g – right. As rounds progress, the correct (yellow) node is found, leading to well converged results similar to if the Stem 3 was initially docked at the yellow node (subsequent panels).
Extended Data Fig. 4 Per-nucleotide modelling convergence of the SARS-CoV-2 Frameshift Stimulation Element (FSE).
a. The same secondary structure as shown in Fig. 2 (without ambiguous basepairing emphasized). Diagrammed here for ease of reading. b. pairwise root mean squared deviation at each nucleotide position over the top 10 autoDRRAFTER derived models, orange line indicates the global mean pairwise root mean squared deviation of 6.84 Å.
Extended Data Fig. 5 De novo models from Rosetta’s rna_denovo application for Frameshift Stimulation Elements from a range of betacoronaviruses, including murine hepatitis virus (MHV), human coronavirus HKU1 (HKU1), human coronavirus OC43 (HCoV-OC43), bovine coronavirus (BCoV), Middle Eastern respiratory syndrome-related coronavirus (MERS), SARS coronavirus (SARS-CoV-1) and SARS-CoV-2.
Top-scoring cluster centers that show a. threaded topologies and b. unthreaded topologies are depicted. Models are only shown if a cluster in the top-scoring 10 clusters shows the desired topology. The 5’ strand of Stem 1 and upstream Slippery Site are colored in dark gray, the 3’ strand of Stem 1 is colored in green, the junction J3/2 is colored in magenta, Stem 2 is colored in blue, and Stem 3 is colored in orange. Yellow circles indicate the ring in each structure.
Extended Data Fig. 6 The secondary structure for the SARS-CoV-2 Frameshift Stimulation Element (FSE), ATP-TTR3 ‘tagged construct’ as determined by 1D chemical SHAPE mapping, as computed in RNAStructure allowing for pseudoknots.
Bootstrapping (100 iterations) support for each continuous helix is shown as an underlined percentage. Nucleotides are colored by a) SHAPE reactivity or b) dimethyl sulfate (DMS) reactivity. Bootstrapping probabilities are shown for the DMS case for helices that are also predicted with RNAstructure guided by DMS reactivity. Nucleotides unreactive to DMS are depicted in grey.
Extended Data Fig. 7 Single-particle cryo-EM analysis of the FSE-ATP-TTR-3.
a. Workflow of cryo-EM data processing of FSE. b. Representative motion-corrected cryo-EM micrograph, randomly selected from the 11,545 micrographs. c. Reference-free 2D class averages. d. Gold standard FSC plots calculated in cryoSPARC. e. Euler angle distribution of the particle images.
Extended Data Fig. 8 Mean in vitro dual luciferase frameshifting assay as a function of LNA challenge as shown as fold-change ± standard deviation.
Values are normalized as described in methods. a, JL4, b, RL4 and the HIV Gag-Pol Frameshift Stimulation Element (FSE, 5.4% ±0.05 total efficiency at 0 µM) used as controls. Fits derived from a standard binding isotherm fit to the data (see Methods) to estimate IC50s vs SARS-CoV-2 (± standard error in stated units; see Methods): c, Slp2 320 nM ± 40; d, S1D-1 130 nM ± 20; e, S1D-2 280 nM ± 70; f, S1D-3 280 nM ± 90; g, S3D-1 500 nM ± 90; h, S3D-2 180 nM ± 20; i, S2D 1.9 µM ± 1.6.
Extended Data Fig. 9 Apoptosis induction and cytotoxicity of FSE-directed LNAs in cells.
a. Induction of caspase-3/7 activation of 0.01, 0.1, or 1 µM of LNAs in duplicate. Data presented as percent caspase expression relative to non-treated, mock control in each cell-line: Huh-7 and Vero E6 cells (N = 2). Camptothecin (Campto) and a caspase-inducing LNA (Pos_LNA), were used as positive controls in a and b. b-c. LNA cytotoxicity evaluated by PrestoBlue cell viability assay 4 days after LNA treatment in b) Huh-7 cells (N = 2) or c) ACE2-A549 cells (N = 4). Results shown as percent viability relative to non-treated, mock control.
Extended Data Fig. 10 LNAs exhibit a dose-dependent effect on viral replication.
A correlation of expected -1 PRF inhibition for a given LNA at either 25 or 100 nM, as computed using the fitting parameters from Supplementary Fig. 4, to observed change in log10(luminescence) of the SARS-CoV-2-nLuc reporter virus following LNA challenge normalized to the Scrambled LNA control (see Methods). Colors as in Fig. 3 and Extended Data Fig. 8. Linear Spearman correlation coefficient plotted R2 = 0.44, P > 0.0025. The concentration of LNA bears a greater impact on viral replication than its identity.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and Tables 1 and 2.
Supplementary Video 1
Cryo-EM map-derived atomic model of the 28-kDa frameshift stimulation element (FSE) from the SARS-CoV-2 RNA genome.
Supplementary Data 1
Statistical source data for Supplementary Fig. 6a.
Supplementary Data 2
Statistical source data for Supplementary Fig. 4a.
Source data
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Zhang, K., Zheludev, I.N., Hagey, R.J. et al. Cryo-EM and antisense targeting of the 28-kDa frameshift stimulation element from the SARS-CoV-2 RNA genome. Nat Struct Mol Biol 28, 747–754 (2021). https://doi.org/10.1038/s41594-021-00653-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-021-00653-y
This article is cited by
-
SARS-CoV-2 biology and host interactions
Nature Reviews Microbiology (2024)
-
Structure, folding and flexibility of co-transcriptional RNA origami
Nature Nanotechnology (2023)
-
Structure genomics of SARS-CoV-2 and its Omicron variant: drug design templates for COVID-19
Acta Pharmacologica Sinica (2022)
-
Secondary structural ensembles of the SARS-CoV-2 RNA genome in infected cells
Nature Communications (2022)
-
Expansion of the RNAStructuromeDB to include secondary structural data spanning the human protein-coding transcriptome
Scientific Reports (2022)