Abstract
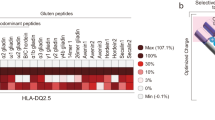
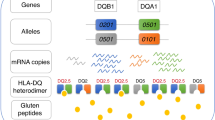
The human leukocyte antigen (HLA) locus is strongly associated with T cell-mediated autoimmune disorders. HLA-DQ2.5-mediated celiac disease (CeD) is triggered by the ingestion of gluten, although the relative roles of genetic and environmental risk factors in CeD is unclear. Here we identify microbially derived mimics of gliadin epitopes and a parental bacterial protein that is naturally processed by antigen-presenting cells and activated gliadin reactive HLA-DQ2.5-restricted T cells derived from CeD patients. Crystal structures of T cell receptors in complex with HLA-DQ2.5 bound to two distinct bacterial peptides demonstrate that molecular mimicry underpins cross-reactivity toward the gliadin epitopes. Accordingly, gliadin reactive T cells involved in CeD pathogenesis cross-react with ubiquitous bacterial peptides, thereby suggesting microbial exposure as a potential environmental factor in CeD.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The structure models and structure factors for the complexes HLA-DQ2.5–P.fluor-α1a, LS2.8/3.15 TCR–HLA-DQ2.5–P.fluor-α1a, and JR5.1 TCR–HLA-DQ2.5–P.aeru-α2a have been deposited at the wwPDB under accession codes PDB 6U3M, 6U3N, and 6U3O, respectively. The source data for Supplementary Figs 1 and 2 are available online.
References
Trowsdale, J. & Knight, J. C. Major histocompatibility complex genomics and human disease. Annu Rev. Genomics Hum. Genet 14, 301–323 (2013).
Dendrou, C. A., Petersen, J., Rossjohn, J. & Fugger, L. HLA variation and disease. Nat. Rev. Immunol. 18, 325–339 (2018).
Pollard, K. M., Hultman, P. & Kono, D. H. Toxicology of autoimmune diseases. Chem. Res. Toxicol. 23, 455–466 (2010).
Marino, E. et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol. 18, 552–562 (2017).
Li, B., Selmi, C., Tang, R., Gershwin, M. E. & Ma, X. The microbiome and autoimmunity: a paradigm from the gut-liver axis. Cell Mol. Immunol. 15, 595–609 (2018).
Sollid, L. M. & Jabri, B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat. Rev. Immunol. 13, 294–302 (2013).
Sollid, L. M., Qiao, S. W., Anderson, R. P., Gianfrani, C. & Koning, F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics 64, 455–460 (2012).
Tye-Din, J. A. et al. Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci. Transl. Med. 2, 41ra51 (2010).
Kim, C. Y., Quarsten, H., Bergseng, E., Khosla, C. & Sollid, L. M. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc. Natl Acad. Sci. USA 101, 4175–4179 (2004).
Henderson, K. N. et al. A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity 27, 23–34 (2007).
Molberg, O. et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 4, 713–717 (1998).
Qiao, S. W. et al. Posttranslational modification of gluten shapes TCR usage in celiac disease. J. Immunol. 187, 3064–3071 (2011).
Broughton, S. E. et al. Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease. Immunity 37, 611–621 (2012).
Petersen, J. et al. T-cell receptor recognition of HLA-DQ2-gliadin complexes associated with celiac disease. Nat. Struct. Mol. Biol. 21, 480–488 (2014).
Qiao, S. W., Christophersen, A., Lundin, K. E. & Sollid, L. M. Biased usage and preferred pairing of α- and β-chains of TCRs specific for an immunodominant gluten epitope in coeliac disease. Int. Immunol. 26, 13–19 (2014).
Vriezinga, S. L. et al. Randomized feeding intervention in infants at high risk for celiac disease. N. Engl. J. Med. 371, 1304–1315 (2014).
Marild, K., Kahrs, C. R., Tapia, G., Stene, L. C. & Stordal, K. Infections and risk of celiac disease in childhood: a prospective nationwide cohort study. Am. J. Gastroenterol. 110, 1475–1484 (2015).
Viitasalo, L. et al. Early microbial markers of celiac disease. J. Clin. Gastroenterol. 48, 620–624 (2014).
Ludvigsson, J. F. & Murray, J. A. Epidemiology of celiac disease. Gastroenterol. Clin. North Am. 48, 1–18 (2019).
Caminero, A. & Verdu, E. F. Celiac disease: should we care about microbes? Am. J. Physiol. Gastrointest. Liver Physiol. 317, G161–G170 (2019).
Verdu, E. F., Galipeau, H. J. & Jabri, B. Novel players in coeliac disease pathogenesis: role of the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 12, 497–506 (2015).
Kagnoff, M. F. et al. Evidence for the role of a human intestinal adenovirus in the pathogenesis of coeliac disease. Gut 28, 995–1001 (1987).
Kagnoff, M. F., Austin, R. K., Hubert, J. J., Bernardin, J. E. & Kasarda, D. D. Possible role for a human adenovirus in the pathogenesis of celiac disease. J. Exp. Med. 160, 1544–1557 (1984).
Dahal-Koirala, S. et al. Discriminative T-cell receptor recognition of highly homologous HLA-DQ2-bound gluten epitopes. J. Biol. Chem. 294, 941–952 (2019).
Purcell, A. W., Ramarathinam, S. H. & Ternette, N. Mass spectrometry-based identification of MHC-bound peptides for immunopeptidomics. Nat. Protoc. 14, 1687–1707 (2019).
Anderson, R. P., Degano, P., Godkin, A. J., Jewell, D. P. & Hill, A. V. S. In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat. Med. 6, 337–342 (2000).
Stene, L. C. et al. Rotavirus infection frequency and risk of celiac disease autoimmunity in early childhood: a longitudinal study. Am. J. Gastroenterol. 101, 2333–2340 (2006).
Bouziat, R. et al. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 356, 44–50 (2017).
Kahrs, C. R. et al. Enterovirus as trigger of coeliac disease: nested case-control study within prospective birth cohort. Br. Med. J. 364, l231 (2019).
Ashorn, S. et al. Elevated serum anti-Saccharomyces cerevisiae, anti-I2 and anti-OmpW antibody levels in patients with suspicion of celiac disease. J. Clin. Immunol. 28, 486–494 (2008).
Riddle, M. S., Murray, J. A. & Porter, C. K. The incidence and risk of celiac disease in a healthy US adult population. Am. J. Gastroenterol. 107, 1248–1255 (2012).
Riddle, M. S., Murray, J. A., Cash, B. D., Pimentel, M. & Porter, C. K. Pathogen-specific risk of celiac disease following bacterial causes of foodborne illness: a retrospective cohort study. Dig. Dis. Sci. 58, 3242–3245 (2013).
Sanchez, E., Donat, E., Ribes-Koninckx, C., Fernandez-Murga, M. L. & Sanz, Y. Duodenal-mucosal bacteria associated with celiac disease in children. Appl Environ. Microbiol. 79, 5472–5479 (2013).
Wacklin, P. et al. Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. Am. J. Gastroenterol. 109, 1933–1941 (2014).
Viitasalo, L. et al. Microbial biomarkers in patients with nonresponsive celiac disease. Dig. Dis. Sci. 63, 3434–3441 (2018).
D’Argenio, V. et al. Metagenomics reveals dysbiosis and a potentially pathogenic N. flavescens strain in duodenum of adult celiac patients. Am. J. Gastroenterol. 111, 879–890 (2016).
Caminero, A. et al. Duodenal bacteria from patients with celiac disease and healthy subjects distinctly affect gluten breakdown and immunogenicity. Gastroenterology 151, 670–683 (2016).
Caminero, A. et al. Duodenal bacterial proteolytic activity determines sensitivity to dietary antigen through protease-activated receptor-2. Nat. Commun. 10, 1198 (2019).
Dickson, R. P. et al. Cell-associated bacteria in the human lung microbiome. Microbiome 2, 28 (2014).
N’Diaye, A. et al. Substance P and calcitonin gene-related peptide: key regulators of cutaneous microbiota homeostasis. Front. Endocrinol. 8, 15 (2017).
Scales, B. S., Dickson, R. P., LiPuma, J. J. & Huffnagle, G. B. Microbiology, genomics, and clinical significance of the Pseudomonas fluorescens species complex, an unappreciated colonizer of humans. Clin. Microbiol. Rev. 27, 927–948 (2014).
Dalwadi, H., Wei, B., Kronenberg, M., Sutton, C. L. & Braun, J. The Crohna’s disease-associated bacterial protein I2 is a novel enteric T cell superantigen. Immunity 15, 149–158 (2001).
Sutton, C. L. et al. Identification of a novel bacterial sequence associated with Crohn’s disease. Gastroenterology 119, 23–31 (2000).
Segal, Y. & Shoenfeld, Y. Vaccine-induced autoimmunity: the role of molecular mimicry and immune crossreaction. Cell Mol. Immunol. 15, 586–594 (2018).
Lang, H. L. et al. A functional and structural basis for TCR cross-reactivity in multiple sclerosis. Nat. Immunol. 3, 940–943 (2002).
Cole, D. K. et al. Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity. J. Clin. Investig. 126, 2191–2204 (2016).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Gras, S. et al. Allelic polymorphism in the T cell receptor and its impact on immune responses. J. Exp. Med. 207, 1555–1567 (2010).
Acknowledgements
We thank the staff at the Australian Synchrotron for assistance with data collection and the staff at the Monash Macromolecular crystallization facility. We thank the CeD volunteers who participated in this study. This work was supported by grants from the National Health and Medical Research Council of Australia (NHMRC, program grant no. APP1113293) and the Australian Research Council (ARC, grant no. CE140100011). F.K. is supported by the collaboration project TIMID (no. LSHM18057-SGF) financed by the PPP allowance made available by Top Sector Life Sciences and Health to Samenwerkende Gezondheidsfondsen (SGF) to stimulate public–private partnerships and co-financing by health foundations that are part of the SGF. A.W.P. is supported by an NHMRC Principal Research Fellowship. J.R. is supported by an ARC Laureate Fellowship.
Author information
Authors and Affiliations
Contributions
J.P., H.H.R., L.C., M.T.T., K.-L.L., Y.K.-W., N.P.C. and M.Y.H. contributed to data generation and analysis. Z.C. and J.M. provided key reagents. R.P.A., A.W.P., J.A.T-D. and F.K. contributed to data analysis and manuscript writing. H.H.R. and J.R. are joint senior and corresponding authors and, with J.P., conceived the study, analyzed data and co-wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
R.P.A. and J.A.T.-D. are inventors of patents owned or licensed by ImmusanT Inc. relating to the diagnostic application of gluten challenge, and use of gluten-derived T cell epitopes for use in therapeutics.
Additional information
Peer review information Inês Chen was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5 and Table 1.
Source data
Source Data
Source data Supplementary Fig. 1
Source Data
Source data Supplementary Fig. 2
Rights and permissions
About this article
Cite this article
Petersen, J., Ciacchi, L., Tran, M.T. et al. T cell receptor cross-reactivity between gliadin and bacterial peptides in celiac disease. Nat Struct Mol Biol 27, 49–61 (2020). https://doi.org/10.1038/s41594-019-0353-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0353-4
This article is cited by
-
Antibiotics in the pathogenesis of diabetes and inflammatory diseases of the gastrointestinal tract
Nature Reviews Gastroenterology & Hepatology (2023)
-
Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease
Nature Communications (2023)
-
Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides
Nature (2022)
-
The double-edged sword of gut bacteria in celiac disease and implications for therapeutic potential
Mucosal Immunology (2022)
-
The enemy within the gut: bacterial pathogens in celiac autoimmunity
Nature Structural & Molecular Biology (2020)