Abstract
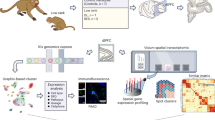
Aging is accompanied by a host of social and biological changes that correlate with behavior, cognitive health and susceptibility to neurodegenerative disease. To understand trajectories of brain aging in a primate, we generated a multiregion bulk (N = 527 samples) and single-nucleus (N = 24 samples) brain transcriptional dataset encompassing 15 brain regions and both sexes in a unique population of free-ranging, behaviorally phenotyped rhesus macaques. We demonstrate that age-related changes in the level and variance of gene expression occur in genes associated with neural functions and neurological diseases, including Alzheimer’s disease. Further, we show that higher social status in females is associated with younger relative transcriptional ages, providing a link between the social environment and aging in the brain. Our findings lend insight into biological mechanisms underlying brain aging in a nonhuman primate model of human behavior, cognition and health.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data generated in this study can be downloaded in raw and processed forms from the NCBI Gene Expression Omnibus under series accession number GSE179330.
Datasets/databases used in this study are publicly available and include the rhesus macaque Mmul_10 reference assembly (NCBI RefSeq accession GCF_003339765.1), Ensembl release 101 (http://aug2020.archive.ensembl.org), KEGG release 100.0 (https://www.genome.jp/kegg), GTEx analysis V8 (dbGaP accession phs000424.v8.p2) and the Alzheimer’s Knowledge Portal AMP-AD meta-analysis (https://doi.org/10.7303/syn11914606).
All requests for biological material should be directed to the CBRU (cbru@pennmedicine.upenn.edu). Requests will be reviewed by the Scientific Review Committee and granted depending on availability, proposed use, investigator funding and other considerations.
Code availability
All code for this study is accessible through the following GitHub repositories: https://github.com/CayoBiobankResearchUnit/brain_transcriptome_aging_bulk (bulk RNA-seq analysis), https://github.com/CayoBiobankResearchUnit/brain_transcriptome_aging_sc (single-nucleus RNA-seq analysis), https://github.com/bbi-lab/bbi-dmux (sci-RNA-seq3 data demultiplexing) and https://github.com/bbi-lab/bbi-sci (sci-RNA-seq3 data preprocessing).
References
Hou, Y. et al. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 15, 565–581 (2019).
Chiou, K. L. et al. Rhesus macaques as a tractable physiological model of human ageing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190612 (2020).
Hernandez-Pacheco, R. et al. Managing the Cayo Santiago rhesus macaque population: the role of density. Am. J. Primatol. 78, 167–181 (2016).
Schneider, J. A., Arvanitakis, Z., Leurgans, S. E. & Bennett, D. A. The neuropathology of probable Alzheimer disease and mild cognitive impairment. Ann. Neurol. 66, 200–208 (2009).
Blair, J. A. et al. Individual case analysis of postmortem interval time on brain tissue preservation. PLoS ONE 11, e0151615 (2016).
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Schaum, N. et al. Ageing hallmarks exhibit organ-specific temporal signatures. Nature 583, 596–602 (2020).
Deleidi, M., Jäggle, M. & Rubino, G. Immune aging, dysmetabolism, and inflammation in neurological diseases. Front. Neurosci. 9, 172 (2015).
Mecocci, P. et al. A long journey into aging, brain aging, and Alzheimer’s disease following the oxidative stress tracks. J. Alzheimers Dis. 62, 1319–1335 (2018).
Sul, J. H., Martin, L. S. & Eskin, E. Population structure in genetic studies: confounding factors and mixed models. PLoS Genet. 14, e1007309 (2018).
Urbut, S. M., Wang, G., Carbonetto, P. & Stephens, M. Flexible statistical methods for estimating and testing effects in genomic studies with multiple conditions. Nat. Genet. 51, 187–195 (2019).
Stephens, M. False discovery rates: a new deal. Biostatistics 18, 275–294 (2017).
Scheff, S. W., Price, D. A., Schmitt, F. A. & Mufson, E. J. Hippocampal synaptic loss in early Alzheimer’s disease and mild cognitive impairment. Neurobiol. Aging 27, 1372–1384 (2006).
Poulin, S. P. et al. Amygdala atrophy is prominent in early Alzheimer’s disease and relates to symptom severity. Psychiatry Res. 194, 7–13 (2011).
Binder, E. B. et al. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat. Genet. 36, 1319–1325 (2004).
Sinclair, D., Fillman, S. G., Webster, M. J. & Weickert, C. S. Dysregulation of glucocorticoid receptor co-factors FKBP5, BAG1 and PTGES3 in prefrontal cortex in psychotic illness. Sci. Rep. 3, 3539 (2013).
Blair, L. J. et al. Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. J. Clin. Invest. 123, 4158–4169 (2013).
Halbgebauer, S. et al. Modified serpinA1 as risk marker for Parkinson’s disease dementia: analysis of baseline data. Sci. Rep. 6, 26145 (2016).
Ebbert, M. T. W. et al. Conserved DNA methylation combined with differential frontal cortex and cerebellar expression distinguishes C9orf72-associated and sporadic ALS, and implicates SERPINA1 in disease. Acta Neuropathol. 134, 715–728 (2017).
Chai, Z., Zheng, P. & Zheng, J. Mechanism of ARPP21 antagonistic intron miR-128 on neurological function repair after stroke. Ann. Clin. Transl. Neurol. 8, 1408–1421 (2021).
Cooper-Knock, J. et al. Mutations in the glycosyltransferase domain of GLT8D1 are associated with familial amyotrophic lateral sclerosis. Cell Rep. 26, 2298–2306 (2019).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Liu, Y., Beyer, A. & Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016).
Greenwood, A. K. et al. The AD Knowledge Portal: a repository for multi-omic data on Alzheimer’s disease and aging. Curr. Protoc. Hum. Genet. 108, e105 (2020).
Izgi, H. et al. Inter-tissue convergence of gene expression during ageing suggests age-related loss of tissue and cellular identity. eLife 11, e68048 (2022).
Holland, P. W. H. & Takahashi, T. The evolution of homeobox genes: implications for the study of brain development. Brain Res. Bull. 66, 484–490 (2005).
Bergsland, M. et al. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 25, 2453–2464 (2011).
Gould, E. How widespread is adult neurogenesis in mammals? Nat. Rev. Neurosci. 8, 481–488 (2007).
Diederich, N. J., James Surmeier, D., Uchihara, T., Grillner, S. & Goetz, C. G. Parkinson’s disease: is it a consequence of human brain evolution? Mov. Disord. 34, 453–459 (2019).
Pouladi, M. A., Morton, A. J. & Hayden, M. R. Choosing an animal model for the study of Huntington’s disease. Nat. Rev. Neurosci. 14, 708–721 (2013).
Finch, C. E. & Austad, S. N. Commentary: is Alzheimer’s disease uniquely human? Neurobiol. Aging 36, 553–555 (2015).
Yerbury, J. J. et al. Walking the tightrope: proteostasis and neurodegenerative disease. J. Neurochem. 137, 489–505 (2016).
Nativio, R. et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat. Neurosci. 21, 497–505 (2018).
Kinney, J. W. et al. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimers Dement. 4, 575–590 (2018).
Vicario-Orri, E., Opazo, C. M. & Muñoz, F. J. The pathophysiology of axonal transport in Alzheimer’s disease. J. Alzheimers Dis. 43, 1097–1113 (2015).
Eschbach, J. & Dupuis, L. Cytoplasmic dynein in neurodegeneration. Pharmacol. Ther. 130, 348–363 (2011).
Glass, C. K., Saijo, K., Winner, B., Marchetto, M. C. & Gage, F. H. Mechanisms underlying inflammation in neurodegeneration. Cell 140, 918–934 (2010).
Wan, Y.-W. et al. Meta-analysis of the Alzheimer’s disease human brain transcriptome and functional dissection in mouse models. Cell Rep. 32, 107908 (2020).
Kumar, S. et al. Extent of dorsolateral prefrontal cortex plasticity and its association with working memory in patients with Alzheimer disease. JAMA Psychiatry 74, 1266–1274 (2017).
Upright, N. A. & Baxter, M. G. Prefrontal cortex and cognitive aging in macaque monkeys. Am. J. Primatol. 83, e23250 (2021).
Prater, K. E. et al. Subtype transcriptomic profiling of myeloid cells in Alzheimer disease brain illustrates the diversity in active microglia phenotypes. Preprint at bioRxiv https://doi.org/10.1101/2021.10.25.465802 (2021).
Bakken, T. E. et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021).
Luebke, J., Barbas, H. & Peters, A. Effects of normal aging on prefrontal area 46 in the rhesus monkey. Brain Res. Rev. 62, 212–232 (2010).
Raible, D. W. & McMorris, F. A. Induction of oligodendrocyte differentiation by activators of adenylate cyclase. J. Neurosci. Res. 27, 43–46 (1990).
Perry, V. H. & Holmes, C. Microglial priming in neurodegenerative disease. Nat. Rev. Neurol. 10, 217–224 (2014).
Niraula, A., Sheridan, J. F. & Godbout, J. P. Microglia priming with aging and stress. Neuropsychopharmacology 42, 318–333 (2017).
Lowsky, D. J., Olshansky, S. J., Bhattacharya, J. & Goldman, D. P. Heterogeneity in healthy aging. J. Gerontol. A Biol. Sci. Med. Sci. 69, 640–649 (2014).
Belsky, D. W. et al. Quantification of biological aging in young adults. Proc. Natl Acad. Sci. USA 112, E4104–E4110 (2015).
Snyder-Mackler, N. et al. Social determinants of health and survival in humans and other animals. Science 368, eaax9553 (2020).
Blomquist, G. E., Sade, D. S. & Berard, J. D. Rank-related fitness differences and their demographic pathways in semi-free-ranging rhesus macaques (Macaca mulatta). Int. J. Primatol. 32, 193–208 (2011).
Snyder-Mackler, N., Somel, M. & Tung, J. Shared signatures of social stress and aging in peripheral blood mononuclear cell gene expression profiles. Aging Cell 13, 954–957 (2014).
Testard, C. et al. Rhesus macaques build new social connections after a natural disaster. Curr. Biol. 31, 2299–2309 (2021).
McColgan, P., Joubert, J., Tabrizi, S. J. & Rees, G. The human motor cortex microcircuit: insights for neurodegenerative disease. Nat. Rev. Neurosci. 21, 401–415 (2020).
Ohm, T. G. The dentate gyrus in Alzheimer’s disease. Prog. Brain Res. 163, 723–740 (2007).
Jiji, S., Smitha, K. A., Gupta, A. K., Pillai, V. P. M. & Jayasree, R. S. Segmentation and volumetric analysis of the caudate nucleus in Alzheimer’s disease. Eur. J. Radiol. 82, 1525–1530 (2013).
Wilson, R. S. et al. Loneliness and risk of Alzheimer disease. Arch. Gen. Psychiatry 64, 234–240 (2007).
Holwerda, T. J. et al. Feelings of loneliness, but not social isolation, predict dementia onset: results from the Amsterdam Study of the Elderly (AMSTEL). J. Neurol. Neurosurg. Psychiatry 85, 135–142 (2014).
Cadar, D. et al. Individual and area-based socioeconomic factors associated with dementia incidence in England: evidence from a 12-year follow-up in the English Longitudinal Study of Ageing. JAMA Psychiatry 75, 723–732 (2018).
Berard, J. A four-year study of the association between male dominance rank, residency status, and reproductive activity in rhesus macaques (Macaca mulatta). Primates 40, 159–175 (1999).
Zannas, A. S. et al. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol. 16, 266 (2015).
Zannas, A. S. Epigenetics as a key link between psychosocial stress and aging: concepts, evidence, mechanisms. Dialogues Clin. Neurosci. 21, 389–396 (2019).
Harvanek, Z. M., Fogelman, N., Xu, K. & Sinha, R. Psychological and biological resilience modulates the effects of stress on epigenetic aging. Transl. Psychiatry 11, 601 (2021).
Miller, G. E., Cohen, S. & Ritchey, A. K. Chronic psychological stress and the regulation of pro-inflammatory cytokines: a glucocorticoid-resistance model. Health Psychol. 21, 531–541 (2002).
Snyder-Mackler, N. et al. Social status alters immune regulation and response to infection in macaques. Science 354, 1041–1045 (2016).
Snyder-Mackler, N. et al. Social status alters chromatin accessibility and the gene regulatory response to glucocorticoid stimulation in rhesus macaques. Proc. Natl Acad. Sci. USA 116, 1219–1228 (2019).
Kessler, M. J. & Rawlins, R. G. A 75-year pictorial history of the Cayo Santiago rhesus monkey colony. Am. J. Primatol. 78, 6–43 (2016).
Missakian, E. A. Genealogical and cross-genealogical dominance relations in a group of free-ranging rhesus monkeys (Macaca mulatta) on Cayo Santiago. Primates 13, 169–180 (1972).
Widdig, A. et al. Low incidence of inbreeding in a long-lived primate population isolated for 75 years. Behav. Ecol. Sociobiol. 71, 18 (2017).
Finch, C. E. & Austad, S. N. Primate aging in the mammalian scheme: the puzzle of extreme variation in brain aging. Age 34, 1075–1091 (2012).
Roth, G. S. et al. Aging in rhesus monkeys: relevance to human health interventions. Science 305, 1423–1426 (2004).
Kessler, M. J., Rawlins, R. G. & London, W. T. The hemogram, serum biochemistry, and electrolyte profile of aged rhesus monkeys (Macaca mulatta). J. Med. Primatol. 12, 184–191 (1983).
Hoffman, C. L., Higham, J. P., Mas-Rivera, A., Ayala, J. E. & Maestripieri, D. Terminal investment and senescence in rhesus macaques (Macaca mulatta) on Cayo Santiago. Behav. Ecol. 21, 972–978 (2010).
Kessler, M. J., Turnquist, J. E., Pritzker, K. P. & London, W. T. Reduction of passive extension and radiographic evidence of degenerative knee joint diseases in cage-raised and free-ranging aged rhesus monkeys (Macaca mulatta). J. Med. Primatol. 15, 1–9 (1986).
Nussey, D. H., Froy, H., Lemaitre, J.-F., Gaillard, J.-M. & Austad, S. N. Senescence in natural populations of animals: widespread evidence and its implications for bio-gerontology. Ageing Res. Rev. 12, 214–225 (2013).
Bronikowski, A. M. et al. Aging in the natural world: comparative data reveal similar mortality patterns across primates. Science 331, 1325–1328 (2011).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2013).
Testard, C. et al. Social connections predict brain structure in a multidimensional free-ranging primate society. Sci. Adv. 8, eabl5794 (2022).
DeCasien, A. R. et al. Evolutionary and biomedical implications of sex differences in the primate brain transcriptome. Preprint at bioRxiv https://doi.org/10.1101/2022.10.03.510711 (2022).
Wong, K.-S. & Pang, H.-M. Simplifying HT RNA quality & quantity analysis. Genet. Eng. Biotechnol. News 33, 17 (2013).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Warren, W. C. et al. Sequence diversity analyses of an improved rhesus macaque genome enhance its biomedical utility. Science 370, eabc6617 (2020).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2015).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
McInnes, L. & Healy, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://doi.org/10.48550/arXiv.1802.03426 (2018).
Schliep, K. P. phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Lipatov, M., Sanjeev, K., Patro, R. & Veeramah, K. Maximum likelihood estimation of biological relatedness from low coverage sequencing data. Preprint at bioRxiv https://doi.org/10.1101/023374 (2015).
Alexa, A., Rahnenführer, J. & Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 (2006).
Alexa, A. & Rahnenführer, J. topGO: enrichment analysis for Gene Ontology. Bioconductor https://doi.org/10.18129/B9.bioc.topGO (2019).
Durinck, S., Spellman, P. T., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57, 289–300 (1995).
Kang, H. M. et al. Efficient control of population structure in model organism association mapping. Genetics 178, 1709–1723 (2008).
Naqvi, S. et al. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science 365, eaaw7317 (2019).
Pletscher-Frankild, S., Pallejà, A., Tsafou, K., Binder, J. X. & Jensen, L. J. DISEASES: text mining and data integration of disease-gene associations. Methods 74, 83–89 (2015).
Wang, M. et al. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci. Data 5, 180185 (2018).
De Jager, P. L. et al. A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci. Data 5, 180142 (2018).
Allen, M. et al. Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci. Data 3, 160089 (2016).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Dunn, P. K. & Smyth, G. K. dglm: double generalized linear models. R package version 1.8.4 https://CRAN.R-project.org/package=dglm (2020).
Herrero, J. et al. Ensembl comparative genomics resources. Database 2016, bav096 (2016).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Cao, J. et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017).
Hennig, B. P. et al. Large-scale low-cost NGS library preparation using a robust Tn5 purification and tagmentation protocol. G3 8, 79–89 (2018).
Korneliussen, T. S. & Moltke, I. NgsRelate: a software tool for estimating pairwise relatedness from next-generation sequencing data. Bioinformatics 31, 4009–4011 (2015).
Hanghøj, K., Moltke, I., Andersen, P. A., Manica, A. & Korneliussen, T. S. Fast and accurate relatedness estimation from high-throughput sequencing data in the presence of inbreeding. Gigascience 8, giz034 (2019).
Korneliussen, T. S., Albrechtsen, A. & Nielsen, R. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014).
Hedrick, P. W. & Lacy, R. C. Measuring relatedness between inbred individuals. J. Hered. 106, 20–25 (2015).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291 (2019).
Darmanis, S. et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl Acad. Sci. USA 112, 7285–7290 (2015).
Zhu, Y. et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, eaat8077 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Tasic, B. et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018).
Miller, J. A. et al. Common cell type nomenclature for the mammalian brain. eLife 9, e59928 (2020).
Yao, Z. et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241 (2021).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019).
Packer, J. S. et al. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science 365, eaax1971 (2019).
McKenzie, A. T. et al. Brain cell type specific gene expression and co-expression network architectures. Sci. Rep. 8, 8868 (2018).
Chen, L. et al. GMPR: a robust normalization method for zero-inflated count data with application to microbiome sequencing data. PeerJ 6, e4600 (2018).
Peters, M. J. et al. The transcriptional landscape of age in human peripheral blood. Nat. Commun. 6, 8570 (2015).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22 (2010).
Anderson, J. A. et al. High social status males experience accelerated epigenetic aging in wild baboons. eLife 10, e66128 (2021).
Acknowledgements
We thank those who make our research possible, particularly the CPRC and the Cayo Santiago Field Station staff, without whom we would not be able to study this amazing system. We also thank the staff of the Sabana Seca Field Station for assistance with sample collection and J. Cao, A. Lea, N. Simons, R. Campbell, J. Tung, I. Schneider-Crease and the University of Washington Basic Biology of Aging training group for valuable feedback at various stages throughout the project. Funding for this research was provided by the National Institutes of Health (R01-MH118203 to M.L.P., U01-MH121260 to N.S.-M., M.L.P., and J. Shendure, R01-MH096875 to M.L.P., R01-AG060931 to N.S.-M., L.J.N.B., and J.P.H., R00-AG051764 to N.S.-M., R01-NS097537 to J.M.N., R35-GM124827 to M.A.W., K99-AG075241 to K.L.C. and P40-OD012217 to M.I.M.), the National Science Foundation (BCS-1800558 to J.P.H. and BCS-1752393 to A.R.D.) and a pilot grant to N.S.-M. from the Brotman Baty Institute. K.L.C. was supported by National Institutes of Health fellowship T32-AG000057 during this research.
The GTEx Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health (https://commonfund.nih.gov/GTEx). Additional funds were provided by the NCI, NHGRI, NHLBI, NIDA, NIMH and NINDS. Donors were enrolled at Biospecimen Source Sites funded by NCI\Leidos Biomedical Research, Inc., subcontracts to the National Disease Research Interchange (10XS170), Roswell Park Cancer Institute (10XS171) and Science Care, Inc. (X10S172). The Laboratory, Data Analysis and Coordinating Center (LDACC) was funded through a contract (HHSN268201000029C) to The Broad Institute, Inc. Biorepository operations were funded through a Leidos Biomedical Research, Inc., subcontract to Van Andel Research Institute (10ST1035). Additional data repository and project management were provided by Leidos Biomedical Research. (HHSN261200800001E). The Brain Bank was supported by supplements to University of Miami grant DA006227. Statistical Methods development grants were made to the University of Geneva (MH090941 and MH101814), the University of Chicago (MH090951, MH090937, MH101825 and MH101820), the University of North Carolina Chapel Hill (MH090936), North Carolina State University (MH101819), Harvard University (MH090948), Stanford University (MH101782), Washington University (MH101810) and the University of Pennsylvania (MH101822). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000424.v8.p2.
The results published here are in whole or in part based on data obtained from the AD Knowledge Portal (https://adknowledgeportal.org/). MayoRNAseq data were provided by the following sources: The Mayo Clinic Alzheimer’s Disease Genetic Studies, led by N. Taner and S. G. Younkin, Mayo Clinic, Jacksonville, FL, using samples from the Mayo Clinic Study of Aging, the Mayo Clinic Alzheimer’s Disease Research Center and the Mayo Clinic Brain Bank. Data collection was supported through funding by NIA grants P50 AG016574, R01 AG032990, U01 AG046139, R01 AG018023, U01 AG006576, U01 AG006786, R01 AG025711, R01 AG017216 and R01 AG003949, NINDS grant R01 NS080820, CurePSP Foundation and support from Mayo Foundation. Study data include samples collected through the Sun Health Research Institute Brain and Body Donation Program of Sun City, AZ. The Brain and Body Donation Program is supported by the NINDS (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the NIA (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research. MSBB data were generated from postmortem brain tissue collected through the Mount Sinai VA Medical Center Brain Bank and were provided by E. Schadt from Mount Sinai School of Medicine. ROSMAP data were provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, IL. Data collection was supported through funding by NIA grants P30AG10161 (ROS), R01AG15819 (ROSMAP; genomics and RNA-seq), R01AG17917 (MAP), R01AG30146, R01AG36042 (5hC methylation and ATACseq), RC2AG036547 (H3K9Ac), R01AG36836 (RNA-seq), R01AG48015 (monocyte RNA-seq) RF1AG57473 (single-nucleus RNA-seq), U01AG32984 (genomics and whole-exome sequencing), U01AG46152 (ROSMAP AMP-AD and targeted proteomics), U01AG46161 (TMT proteomics), U01AG61356 (whole-genome sequencing, targeted proteomics and ROSMAP AMP-AD), the Illinois Department of Public Health (ROSMAP) and the Translational Genomics Research Institute (genomics). Additional phenotypic data can be requested at www.radc.rush.edu.
Author information
Authors and Affiliations
Consortia
Contributions
N.S.-M., M.L.P., M.J.M., J.P.H., L.J.N.B., K.L.C. and A.R.D. conceptualized the research. N.S.-M., J. Shendure, M.L.P., M.J.M., L.M.S. and K.L.C. conceptualized the single-cell sequencing studies. M.J.M., N.S.-M., K.L.C., A.R.D., O.G., N.R.C., S.E.B.S., M.C.J., C.J.W. and S.T. collected brain tissue, facilitated by M.I.M., A.V.R.-L., J. Sallet, C.S.W., S.C.A., M.K.S., A.D.M., J.P.H., M.L.P. and CBRU. M.A.W. contributed data. K.L.C., A.R.D., A.M. and C.H.S. performed genomic lab work. J.E.N.-D.V. collected behavioral data using a protocol designed by L.J.N.B. K.L.C., A.R.D., C.H.S., A.A.G. and H.A.P. performed genomic analysis with input from N.S.-M. and J. Shendure. C.T. and L.J.N.B. performed behavioral analysis. K.P.R. and J.M.N. performed protein experiments and analysis. K.L.C., A.R.D. and N.S.-M. wrote the manuscript. All authors reviewed and revised the manuscript. Full membership of the CBRU: S. C. Antón, L. J. N. Brent, J. P. Higham, M. I. Martínez, A. D. Melin, M. J. Montague, M. L. Platt, J. Sallet and N. Snyder-Mackler
Corresponding authors
Ethics declarations
Competing interests
J. Shendure is a scientific advisory board member, consultant and/or cofounder of Cajal Neuroscience, Guardant Health, Maze Therapeutics, Camp4 Therapeutics, Phase Genomics, Adaptive Biotechnologies, Scale Biosciences and Sixth Street Capital. M.L.P. is a scientific advisory board member, consultant and/or cofounder of Blue Horizons International, NeuroFlow, Amplio, Cogwear Technologies, Burgeon Labs and Ashurst Cognitive Health and receives research funding from AIIR Consulting, the SEB Group, Mars Inc., Slalom Inc., the Lefkort Family Research Foundation, Sisu Capital and Benjamin Franklin Technology Partners. All other authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Sample and study design.
(a) Location of Cayo Santiago off the southeastern coast of Puerto Rico. (b) Age distribution of individuals sampled for the bulk tissue RNA sequencing dataset. Selected individuals span the natural adult age distribution and are evenly balanced between sexes. (c) Age distribution of individuals per brain region (excludes samples not passing laboratory or bioinformatic quality control). (d) Age distribution of females sampled for the single-nucleus RNA sequencing dataset. Selected individuals span the natural age distribution and are evenly balanced between two naturally occurring social groups (HH and KK) on Cayo Santiago. (e) Hazard rates from a previously published demographic model of Cayo Santiago macaques (ref. 2) demonstrate that individuals in the age range of our sample experience age-associated increases in mortality risk (N = 11,659 biologically independent animals). Error bars represent the mean hazard rate ± the standard error.
Extended Data Fig. 2 Global similarity of gene expression across brain regions.
(a) Dendrogram showing hierarchical clustering results on averaged expression for each brain region. (b) The same dendrogram visualized over 1000 bootstrap replicates in which libraries were randomly sampled with replacement before calculating average expression and repeating hierarchical clustering. (c) Dendrogram showing hierarchical clustering results on all libraries. Terminal branches are colored by brain region.
Extended Data Fig. 3 Comparison of variance partitioning of age between macaques and humans.
(a) Age explains similar proportions of variance in global gene expression across diverse brain regions between macaques from this study (left) and humans from the GTEx study (right). Median variance explained by age was 0.19% (interquartile range [IQR] = 0.04–0.51%) in this study (N = 527 biologically independent samples) and 0.26% (IQR = 0.07–0.82%) in the GTEx study (N = 2,642 biologically independent samples). Effects of technical covariates were first removed to facilitate this comparison. (b) Age explains similar proportions of variance in gene expression within individual brain regions. Median variance explained by age ranged from 0.6–6.4% across brain regions from this study (N = 36 biologically independent animals, left) and ranged from 0.3–4.2% across brain regions from the GTEx study (N = 382 biologically independent individuals, right). Box plots depict the median (center), and IQR (bounds of box), with whiskers extending to either the maxima/minima or to the median ±1.5×IQR, whichever is nearest.
Extended Data Fig. 4 Analysis of age-associated effects on gene expression levels.
(a) Quantile-quantile (QQ) plots of P values from initial efficient mixed model association (EMMA) tests show enrichment of low P values across nearly all brain regions tested (all EMMA tests are two-sided). (b) A multivariate adaptive shrinkage (MASH) approach substantially improves statistical power by leveraging shared patterns between tissue datasets, resulting in a greater number of genes passing a threshold (local false sign rate [LFSR] < 0.2) relative to a similar threshold applied to our EMMA results (false discovery rate [FDR] < 0.2). (c) The number of significant genes visualized over a range of LFSR thresholds shows extremely stable rank order of brain regions. (d) Whole-brain age-differentially expressed genes (wbaDEGs) cross-referenced with our gene trajectory results (Fig. 1d) demonstrate that the vast majority of wbaDEGs fall into four clusters, marked by asterisks (***). Most wbaDEGs decreasing in expression with age fall into trajectories associated with signaling-related functions, while most wbaDEGs increasing in expression with age fall into a trajectory associated with the immune response. In the bottom panel, the percentage of genes assigned to wbaDEGs is plotted on the y axis.
Extended Data Fig. 5 Similarities of age effects across brain regions.
(a) Proportion of aDEGs with shared magnitude between brain regions. aDEGs share magnitude between regions when they share signs and their effect estimates are within a factor of 2 from one another. These conditions for sharing are more stringent than those in Fig. 2b, which does not require the latter criterion. For each pair of regions, genes are included if they were significant (LFSR < 0.2) in either region. (b) Upset plot highlights brain region combinations with the greatest number of aDEGs with shared signs. Note that among the top region combinations are single-region-specific aDEGs as well as wbaDEGs. (c) Proportion of aDEGs that exhibit (top) shared signs or (bottom) shared magnitudes across variable numbers of brain regions. These distributions reveal that subcortical regions sampled exhibited slightly broader sharing than cortical regions. For this analysis, genes are included if they were significant (LFSR < 0.2) in any region.
Extended Data Fig. 6 Cell-type marker genes and proportional changes with age.
(a) Expression of top 5 marker genes (according to the pseudo-R2 statistic) for 8 assigned cell types. (b) Age plotted against cell cluster percentages. Asterisks in cluster titles indicate a significant effect of age on cell cluster percentages based on a linear model (two-sided test, excitatory neurons 6: Bonferroni-adjusted P = 0.014; excitatory neurons 10: Bonferroni-adjusted P = 0.024).
Extended Data Fig. 7 Age-associated differences in gene expression within single cell types.
(a) QQ plot of EMMA tests on pseudobulk data show a strong enrichment of low P values (all EMMA tests are two-sided). (b) Comparison of model estimates from EMMA (FDR < 0.2) and MASH (LFSR < 0.2) show similar numbers of significant genes. (c). Cell-type aDEG counts (MASH) classified according to overlap with expressed genes and aDEGs from bulk RNA-seq analysis of the dlPFC. (d). Percentages of aDEGs overlapping with aDEGs from bulk RNA-seq analysis of the dlPFC (excluding genes not expressed in both modalities). (e) Rank order of significant genes per cell type are robust across a range of significance (LFSR) thresholds. (f) Upset plot shows rank order of cell type combinations with the greatest number of genes exhibiting significant age-associated differences in expression.
Extended Data Fig. 8 Cell-type deconvolution of bulk tissue RNA-seq data.
(a) After adjusting for cell type composition and repeating our EMMA and MASH models, similar numbers of genes per brain region pass our significance thresholds (LFSR < 0.2). (b) Standardized age effect estimates from analyses controlling and not controlling for cell-type composition are strongly positively correlated, but slopes < 1 in most brain regions suggest that some age effects identified in our bulk gene expression results are due to age-related changes in cell proportion. (c) The majority of aDEGs belonging to four clusters with pronounced age-associated directional changes (Fig. 1d) met our criteria (LFSR < 0.2) as aDEGs after controlling for heterogeneous tissue compositions using cell-type deconvolution (CD) analysis. Substantial fractions, however, did not, providing further evidence that some results at the bulk-tissue level are driven by age-related changes in cell proportion.
Extended Data Fig. 9 Exploratory analyses of single-cell-type pseudobulk data from the dlPFC.
UMAP plots of pseudobulk libraries reveal that the latent structure is driven primarily by (a) cell type and not (b) age. (c) Single-nucleus libraries were aggregated across all cell types to approximate bulk tissue RNA sequencing libraries. Age effects from pseudobulk dlPFC libraries were then compared to age effects from bulk dlPFC libraries, showing a strong positive correlation. Colors denote whether age effects were considered significant for both datasets (EMMA test uncorrected P < 0.05). Error bands represent the 95% confidence interval of linear model predictions. (d) Similarly, age prediction on pseudobulk libraries using the wbaDEGs model (see Methods) developed from analysis of bulk RNA-seq data shows a similar positive correlation (linear model statistics are presented) and accuracy to predictions on bulk tissue libraries. Error bands represent the 95% confidence interval of linear model predictions.
Supplementary information
Supplementary Information
Supplementary Figs. 1–17.
Supplementary Table
Supplementary Tables 1–37.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chiou, K.L., DeCasien, A.R., Rees, K.P. et al. Multiregion transcriptomic profiling of the primate brain reveals signatures of aging and the social environment. Nat Neurosci 25, 1714–1723 (2022). https://doi.org/10.1038/s41593-022-01197-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-022-01197-0