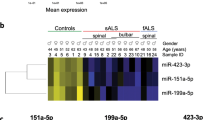
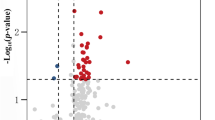
Abstract
Amyotrophic lateral sclerosis (ALS) is a relentless neurodegenerative disease of the human motor neuron system, where variability in progression rate limits clinical trial efficacy. Therefore, better prognostication will facilitate therapeutic progress. In this study, we investigated the potential of plasma cell-free microRNAs (miRNAs) as ALS prognostication biomarkers in 252 patients with detailed clinical phenotyping. First, we identified, in a longitudinal cohort, miRNAs whose plasma levels remain stable over the course of disease. Next, we showed that high levels of miR-181, a miRNA enriched in neurons, predicts a greater than two-fold risk of death in independent discovery and replication cohorts (126 and 122 patients, respectively). miR-181 performance is similar to neurofilament light chain (NfL), and when combined together, miR-181 + NfL establish a novel RNA–protein biomarker pair with superior prognostication capacity. Therefore, plasma miR-181 alone and a novel miRNA–protein biomarker approach, based on miR-181 + NfL, boost precision of patient stratification. miR-181-based ALS biomarkers encourage additional validation and might enhance the power of clinical trials.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Source data for figures are provided in supplementary tables. FASTQ.gz files with raw sequencing data, text files with raw read counts, Excel files with processed read counts and R codes are available as GSE168714 in the Gene Expression Omnibus. Source data are provided with this paper.
References
Munsat, T. L., Andres, P. L., Finison, L., Conlon, T. & Thibodeau, L. The natural history of motoneuron loss in amyotrophic lateral sclerosis. Neurology 38, 409–413 (1988).
Ravits, J. M. & La Spada, A. R. ALS motor phenotype heterogeneity, focality, and spread: deconstructing motor neuron degeneration. Neurology 73, 805–811 (2009).
Benatar, M. et al. ALS biomarkers for therapy development: state of the field and future directions. Muscle Nerve 53, 169–182 (2016).
Chipika, R. H., Finegan, E., Li Hi Shing, S., Hardiman, O. & Bede, P. Tracking a fast-moving disease: longitudinal markers, monitoring, and clinical trial endpoints in ALS. Front. Neurol. 10, 229 (2019).
Verber, N. S. et al. Biomarkers in motor neuron disease: a state of the art review. Front. Neurol. 10, 291 (2019).
Rossi, D. et al. CSF neurofilament proteins as diagnostic and prognostic biomarkers for amyotrophic lateral sclerosis. J. Neurol. 265, 510–521 (2018).
Goncalves, M. et al. Phosphoneurofilament heavy chain and vascular endothelial growth factor as cerebrospinal fluid biomarkers for ALS. Amyotroph. Lateral Scler. Frontotemporal Degener. 18, 134–136 (2017).
Lu, C. H. et al. Plasma neurofilament heavy chain levels and disease progression in amyotrophic lateral sclerosis: insights from a longitudinal study. J. Neurol. Neurosurg. Psychiatry 86, 565–573 (2015).
Gonzalez-Garza, M. T., Martinez, H. R., Cruz-Vega, D. E., Hernandez-Torre, M. & Moreno-Cuevas, J. E. Adipsin, MIP-1b, and IL-8 as CSF biomarker panels for ALS diagnosis. Dis. Markers 2018, 3023826 (2018).
Prado, L. G. R. et al. Longitudinal assessment of clinical and inflammatory markers in patients with amyotrophic lateral sclerosis. J. Neurol. Sci. 394, 69–74 (2018).
Lu, C. H. et al. Systemic inflammatory response and neuromuscular involvement in amyotrophic lateral sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 3, e244 (2016).
Haramati, S. et al. miRNA malfunction causes spinal motor neuron disease. Proc. Natl Acad. Sci. USA 107, 13111–13116 (2010).
Emde, A. et al. Dysregulated miRNA biogenesis downstream of cellular stress and ALS-causing mutations: a new mechanism for ALS. EMBO J. 34, 2633–2651 (2015).
Eitan, C. & Hornstein, E. Vulnerability of microRNA biogenesis in FTD–ALS. Brain Res. 1647, 105–111 (2016).
Takahashi, I. et al. Identification of plasma microRNAs as a biomarker of sporadic amyotrophic lateral sclerosis. Mol. Brain 8, 67 (2015).
de Andrade, H. M. et al. MicroRNAs-424 and 206 are potential prognostic markers in spinal onset amyotrophic lateral sclerosis. J. Neurol. Sci. 368, 19–24 (2016).
Sheinerman, K. S. et al. Circulating brain-enriched microRNAs as novel biomarkers for detection and differentiation of neurodegenerative diseases. Alzheimers Res. Ther. 9, 89 (2017).
Dobrowolny, G. et al. A longitudinal study defined circulating microRNAs as reliable biomarkers for disease prognosis and progression in ALS human patients. Cell Death Discov. 7, 4 (2021).
Benigni, M. et al. Identification of miRNAs as potential biomarkers in cerebrospinal fluid from amyotrophic lateral sclerosis patients. Neuromolecular Med. 18, 551–560 (2016).
Coenen-Stass, A. M. L. et al. Evaluation of methodologies for microRNA biomarker detection by next generation sequencing. RNA Biol. 15, 1133–1145 (2018).
Ogłuszka, M., Orzechowska, M., Jędroszka, D., Witas, P. & Bednarek, A. K. Evaluate Cutpoints: adaptable continuous data distribution system for determining survival in Kaplan–Meier estimator. Comput. Methods Prog. Biomed. 177, 133–139 (2019).
Austin, P. C. & Tu, J. V. Bootstrap methods for developing predictive models. Am. Statistician 58, 131–137 (2004).
Westeneng, H. J. et al. Prognosis for patients with amyotrophic lateral sclerosis: development and validation of a personalised prediction model. Lancet Neurol. 17, 423–433 (2018).
Steinbach, R. et al. Disease aggressiveness signatures of amyotrophic lateral sclerosis in white matter tracts revealed by the D50 disease progression model. Hum. Brain Mapp. 42, 737–752 (2021).
Reichenstein, I. et al. Human genetics and neuropathology suggest a link between miR-218 and amyotrophic lateral sclerosis pathophysiology. Sci. Transl. Med. 11, eaav5264 (2019).
Ludwig, N. et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 44, 3865–3877 (2016).
Khalil, M. et al. Neurofilaments as biomarkers in neurological disorders. Nat. Rev. Neurol. 14, 577–589 (2018).
Lu, C. H. et al. Neurofilament light chain: a prognostic biomarker in amyotrophic lateral sclerosis. Neurology 84, 2247–2257 (2015).
Poesen, K. & Van Damme, P. Diagnostic and prognostic performance of neurofilaments in ALS. Front. Neurol. 9, 1167 (2018).
Benatar, M. et al. Validation of serum neurofilaments as prognostic and potential pharmacodynamic biomarkers for ALS. Neurology 95, e59–e69 (2020).
Corradi, E. et al. Axonal precursor miRNAs hitchhike on endosomes and locally regulate the development of neural circuits. EMBO J. 39, e102513 (2020).
Chen, C.-Z., Li, L., Lodish, H. F. & Bartel, D. P. MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83 (2004).
Benatar, M., Wuu, J., Andersen, P. M., Lombardi, V. & Malaspina, A. Neurofilament light: a candidate biomarker of presymptomatic amyotrophic lateral sclerosis and phenoconversion. Ann. Neurol. 84, 130–139 (2018).
Puentes, F. et al. Immune reactivity to neurofilaments and dipeptide repeats in ALS progression. Preprint at https://www.biorxiv.org/content/10.1101/2020.02.25.965236v1 (2020).
Puentes, F. et al. Immune reactivity to neurofilament proteins in the clinical staging of amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 85, 274–278 (2014).
Brooks, B. R., Miller, R. G., Swash, M., Munsat, T. L. & World Federation of Neurology Research Group on Motor Neuron Diseases. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 1, 293–299 (2000).
Cedarbaum, J. M. et al. The ALSFRS-R: a revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase III). J. Neurol. Sci. 169, 13–21 (1999).
Poesen, K. et al. Neurofilament markers for ALS correlate with extent of upper and lower motor neuron disease. Neurology 88, 2302–2309 (2017).
Prell, T. et al. Reaction to endoplasmic reticulum stress via ATF6 in amyotrophic lateral sclerosis deteriorates with aging. Front. Aging Neurosci. 11, 5 (2019).
Kohen, R. et al. UTAP: user-friendly transcriptome analysis pipeline. BMC Bioinformatics 24, 154 (2019).
Kozomara, A. & Griffiths-Jones, S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Lombardi, V. et al. Muscle and not neuronal biomarkers correlate with severity in spinal and bulbar muscular atrophy. Neurology 92, e1205–e1211 (2019).
Peng, T. et al. A BaSiC tool for background and shading correction of optical microscopy images. Nat. Commun. 8, 14836 (2017).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Benjamini, Y., Drai, D., Elmer, G., Kafkafi, N. & Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284 (2001).
Harrell, F. E. Jr., Lee, K. L., Califf, R. M., Pryor, D. B. & Rosati, R. A. Regression modelling strategies for improved prognostic prediction. Stat. Med. 3, 143–152 (1984).
Grubbs, F. E. Procedures for detecting outlying observations in samples. Technometrics 11, 1–21 (1969).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2015).
Acknowledgements
We thank V. Lombardi (University College London) for technical assistance and I. Ben-Dov (Hadassah Hebrew University Medical Center) for advice on statistics. We acknowledge patients for their contributions and all ALS biomarker study coworkers and their contributions to the biobanking project, which made this study possible (REC 09/H0703/27). We also thank the North Thames Local Research Network for its support and life science editors for editorial assistance. E.H. is the Mondry Family Professorial Chair and Head of the Nella and Leon Benoziyo Center for Neurological Diseases. Imaging was performed at the de Picciotto Cancer Cell Observatory, in memory of Wolfgang and Ruth Lesser. Funding: this research was supported by the following grants: Motor Neurone Disease Association (MNDA no. 839-791), Redhill Foundation – Sam and Jean Rothberg Charitable Trust and J. and E. Moravitz. Research at the Hornstein lab is supported by the CReATe Consortium and ALSA (program: ‘Prognostic value of miRNAs in biofluids from ALS patients’); the RADALA Foundation; AFM Telethon (20576); Weizmann - Brazil Center for Research on Neurodegeneration at the Weizmann Institute of Science; the Minerva Foundation, with funding from the Federal German Ministry for Education and Research; the ISF Legacy Heritage Fund 828/17; the Israel Science Foundation 135/16 and ISF IPMP 3497/21; Target ALS 118945; the Thierry Latran Foundation for ALS Research; the European Research Council, under the European Union’s Seventh Framework Programme (FP7/2007–2013)/ERC grant agreement no. 617351; ERA-Net for Research Programmes on Rare Diseases (eRARE FP7), via the Israel Ministry of Health; Dr. Sydney Brenner and friends; A. Alfred Taubman through IsrALS; Yeda-Sela; Yeda-CEO; the Israel Ministry of Trade and Industry; the Y. Leon Benoziyo Institute for Molecular Medicine; the Kekst Family Institute for Medical Genetics; the David and Fela Shapell Family Center for Genetic Disorders Research; the Crown Human Genome Center; the Nathan, Shirley, Philip and Charlene Vener New Scientist Fund; the Julius and Ray Charlestein Foundation; the Fraida Foundation; the Wolfson Family Charitable Trust; the Adelis Foundation; Merck (United Kingdom); M. Halphen; and the estates of F. Sherr, L. Asseof and L. Fulop. P.F. is supported by a Medical Research Council Senior Clinical Fellowship and the Lady Edith Wolfson Fellowship scheme (MR/M008606/1 and MR/S006508/1). J.G. was supported in the JPND framework ONWebDUALS, and L.G. is the Graeme Watts Senior Research Fellow supported by the Brain Research Trust. N.S.Y. was supported by the Israeli Council for Higher Education via the Weizmann Data Science Research Center, by a research grant from the Estate of Tully and Michele Plesser and by Maccabim Foundation. I.M. was supported by Teva Pharmaceutical Industries as part of the Israeli National Network of Excellence in Neuroscience (fellowship no. 117941).
Author information
Authors and Affiliations
Contributions
I.M., P.F., A.M. and E.H. conceived the research. I.M., N.S.Y., J.G. and E.H. analyzed the data. A.M. and C.H.L. established cohorts, obtained ethics approval and collected human samples for research. E.Y. performed in situ hybridization. A.C.S. assisted with research. I.M., N.S.Y., P.F., A.M. and E.H. wrote the manuscript, with comments from and final approval by all other authors. L.G. provided resources for research and input in research development. A.M. is the corresponding author for cohorts and clinical data. P.F. and E.H. are corresponding authors for all other facets of the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Analysis of miRNAs that increase during ALS course.
(a) Plasma levels of four miRNAs of the 129 miRNAs analyzed in main Fig. 1a displayed low inter-individual variability, but increased with the disease course, suggesting that, although they are not suited for prognostic use, they could potentially monitor disease progression. (miR-423/484/92a/b, t4/t1 > 1.5 SD, X-axis) (B-D) MA plots of differential miRNA expression upon repeated sampling relative to the first phlebotomy. Red features denote miRNAs with statistically significant change in levels. Temporal changes in the levels of (E) miR-423-5p, (F) miR-484, (G) miR-92a-3p, or (H) miR-92b-3p revealed correlation with time passing from enrollment (in months). Spaghetti plots of individual patient trajectories (t1-t4) denoted for (I) miR-423-5p, t21 = 4.52, p = 0.0002; (J) miR-484, t21 = 4.05,p = 0.0006; (K) miR-92a-3p, t21 = 3.85, p = 0.0009 or (L) miR-92b-3p, t21 = 4.77, p = 0.0001. Time intervals: t1-t2 6.3 ± 0.3 m.; t1-t3 13.0 ± 0.3 m.; t1-t4 32.7 ± 3 m. Disease duration: t1 28.8 ± 3 m.; t4 61.5 ± 3 m. Validation of changes to miRNA levels in an independent replication cohort (N = 26 individuals, Table 2). Spaghetti plots of individual patient trajectories (t1-t2 13.7 ± 1.6 months) in a replication cohort, for (M) miR-423-5p, t24 = 0.98, p = 0.17 (N) miR-484, t25 = 2.08, p = 0.02; (O) miR-92a-3p, t25 = 2.13, p = 0.02; or (P) miR-92b-3p, t23 = 1.55, p = 0.067. Together, miR-484 and miR-92a/b may be considered as candidate molecular biomarkers of functional decline over the course of disease. Data presented as Mean ± SEM. *p < 0.05, ***p < 0.001, paired t-test. Analysis of a single miR-423-5p sample and two miR-92b-3p samples in the replication cohort deviated from the mean according to Grubbs test and these were excluded as outliers. Correlation between the relative disease covered (rD50) in longitudinal plasma collections (X-axis) and levels of (Q) miR-423-5p, (R) miR-484, (S) miR-92a-3p, and (T) miR-92b-3p (Y-axis). The relative D50 (rD50) is a derivative of ALS Functional Rating Scale-Revised (ALSFRS-R) decay that reveals the disease covered by individual patients independent of the rate of progression24,39. For example, an rD50 of 0.0 signifies ALS onset, and 0.5 signifies the time-point where functionality is reduced by half. Longitudinal miR-484/92a/b levels in blood correlated with rD50 at the time of sampling (R-T). All statistical tests were two-sided, except for panels M-P.
Extended Data Fig. 2 Clinical features are comparable between discovery and replication cohorts.
(A) Survival from enrollment (B) survival from symptom onset (C) ALSFRS-R score at enrollment (D) progression rate at enrollment (E) sex distribution (F) onset site distribution (G) Riluzole treatment status (H) number of censored patients. Box plots in A-D are presented as median bound between minimum and maximum values. All statistical tests were two-sided. n = 252 biologically independent human samples.
Extended Data Fig. 3 Pipeline for selecting miRNAs as candidate prognostic markers.
2008 miRNAs were aligned to the human genome in the longitudinal study and out of them, 187 miRNAs, which exhibited >50 UMI counts in 60% of the samples, were included in further analysis. 125 out of the 187 miRNAs were longitudinally stable with low interindividual variability (green features in Fig. 1a). In the discovery cohort, 106 out of these 125 miRNAs passed a filtering criterion of average UMI counts >100 across all samples, and were analyzed for prognosis differences between low and high level in the discovery cohort. 19 miRNAs were further excluded after additional QC based on logrank analysis (opposite directions of prognosis differences between members of the same miRNA family, for example miR-27a and miR-27b), and the remaining 87 miRNAs were assessed for logrank and p values for prognosis differences as demonstrated in Fig. 2. 9 out of these 87 miRNAs displayed logrank p ≤ 0.01, and all of their possible pairs (9*8/2 = 36), derived from multiplication of the levels of two single miRNAs, were further assessed for prognosis differences. Nine single miRNAs and 20 miRNA pairs, each displaying a logrank p ≤ 0.01, were further subjected to feature selection by bootstrap resampling. In bootstrap resampling, features had to be selected >70% of the bootstrap samples and display statistical significance in >85% of the samples in which they were selected, in order to be tested as a prognostic marker on the replication cohort. miR-181 was the only feature fulfilling those criteria, hence it was tested in the discovery cohort, and exhibited significant survival differences and hazard ratios, both in the discovery cohort and when validated on a replication cohort that was set aside until that point.
Extended Data Fig. 4 Scatter plot, assessing agreement between separation of survival by 123 miRNA features.
The optimal threshold was calculated per miRNA in a discovery cohort of 126 patients by21. Single miRNAs (black) or miRNA pairs (green), displaying a p-value ≤0.01 (log 10 transformed values ≥ 2), for logrank test from study enrollment (logrank χ2, y-axis) or first symptoms (onset, logrank χ2, x-axis). gray: insignificant features.
Extended Data Fig. 5 miR-181 levels are predictive of survival length regardless of Riluzole treatment.
Survival by miR-181 levels in untreated patients, from enrollment (A) or onset (B), and in patients that were treated with Riluzole, from enrollment (C) or onset (D). All statistical test were two-sided. N = 248 biologically independent human samples.
Extended Data Fig. 6 miR-181 levels in expression bins.
miR-181 levels are higher in the high vs low expression bin, in both the (A) discovery cohort, t21 = 5.94, p < 0.0001 and (B) replication cohort, t21 = 2.87, p = 0.009. Plots depicting inverse correlation between miR-181 levels and survival from first phlebotomy (C), or from disease onset (D). Box plots in A,B are presented as median bound between minimum and maximum values. **p < 0.01, ***p < 0.0001, unpaired t-test with Welch’s correction. All statistical tests were two-sided. N = 248 biologically independent human samples.
Extended Data Fig. 7 miR-181 levels with respect to parameters of the D50 model.
(A) D50, a measure of disease aggressiveness, is significantly lower in high vs low miR-181 levels, indicating a more aggressive disease. t194 = 3.99, p < 0.0001. (B) Individual disease covered, reflected by rD50 values, is not different between low and high miR-181 expression bins. t72 = 1.85, p = 0.07 (C) No correlation of miR-181 levels with individual disease covered. (D) miR-181 levels are not different between different phases of disease defined by rD50 values. One-way ANOVA: F2 = 1.94, p = 0.15. Box plots in A, B and D are presented as median bound between minimum and maximum values. ***p < 0.0001, unpaired t-test with Welch’s correction. All statistical tests were two-sided. N = 248 biologically independent human samples.
Extended Data Fig. 8 miR-181 levels are not related to phenotypic properties at enrollment.
Lack of correlation between miR-181 levels at enrollment and progression rate (A), ALSFRS-R (B) and age at onset (C) in the discovery cohort, or in the replication cohort (D-F). These properties were not different between low and high miR-181 bins (G-I). Box plots in G-I are presented as median bound between minimum and maximum values All statistical test were two-sided. No adjustment for multiple comparisons was performed. N = 248 biologically independent human samples.
Extended Data Fig. 9 Cox proportional hazard analysis for continuous miR-181 and NfL values.
Multivariate Cox proportional hazard analysis for z-scores of miR-181 and NfL from enrollment (A) or onset (B) on the discovery and replication cohorts. Univariate Cox on the merged cohort (discovery + replication) for the z-scores of miR-181, NfL and the sum of the z-scores of both, from enrollment (C) or onset (D). Data are presented as median ± 95% CI. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Wald test.
Extended Data Fig. 10 Association of miR-181 with other markers.
miR-181 is not correlated with NfL levels, either in the full cohort (A) or when NfL is broken into tertiles (B). (C-E) miR-181 is not correlated with markers of muscle integrity (CK and creatinine) or inflammatory marker (TNF-alpha). All statistical test were two-sided. No adjustment for multiple comparisons was performed. N = 248 biologically independent human samples.
Supplementary information
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 7
Statistical Source Data
Source Data Extended Data Fig. 8
Statistical Source Data
Source Data Extended Data Fig. 9
Statistical Source Data
Source Data Extended Data Fig. 10
Statistical Source Data
Rights and permissions
About this article
Cite this article
Magen, I., Yacovzada, N.S., Yanowski, E. et al. Circulating miR-181 is a prognostic biomarker for amyotrophic lateral sclerosis. Nat Neurosci 24, 1534–1541 (2021). https://doi.org/10.1038/s41593-021-00936-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-021-00936-z
This article is cited by
-
Non-coding RNAs in disease: from mechanisms to therapeutics
Nature Reviews Genetics (2024)
-
Opinion: more mouse models and more translation needed for ALS
Molecular Neurodegeneration (2023)
-
Cardiovascular complications of diabetes: role of non-coding RNAs in the crosstalk between immune and cardiovascular systems
Cardiovascular Diabetology (2023)
-
A distinct circular DNA profile intersects with proteome changes in the genotoxic stress-related hSOD1G93A model of ALS
Cell & Bioscience (2023)
-
Amyotrophic lateral sclerosis: a neurodegenerative disorder poised for successful therapeutic translation
Nature Reviews Drug Discovery (2023)