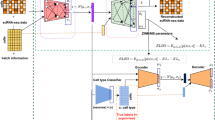
Abstract
Generative pretrained models have achieved remarkable success in various domains such as language and computer vision. Specifically, the combination of large-scale diverse datasets and pretrained transformers has emerged as a promising approach for developing foundation models. Drawing parallels between language and cellular biology (in which texts comprise words; similarly, cells are defined by genes), our study probes the applicability of foundation models to advance cellular biology and genetic research. Using burgeoning single-cell sequencing data, we have constructed a foundation model for single-cell biology, scGPT, based on a generative pretrained transformer across a repository of over 33 million cells. Our findings illustrate that scGPT effectively distills critical biological insights concerning genes and cells. Through further adaptation of transfer learning, scGPT can be optimized to achieve superior performance across diverse downstream applications. This includes tasks such as cell type annotation, multi-batch integration, multi-omic integration, perturbation response prediction and gene network inference.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All sources of used datasets have been reported in Datasets. Pretraining datasets can be retrieved from the CELLxGENE census, release version 15 May 2023 (https://chanzuckerberg.github.io/cellxgene-census/python-api.html, https://cellxgene.cziscience.com/). For the annotation task, the MS dataset was accessed from https://www.ebi.ac.uk/gxa/sc/experiments/E-HCAD-35. The myeloid dataset is publicly accessible from the GEO database using accession number GSE154763. The processed human pancreas dataset was retrieved from https://github.com/JackieHanLab/TOSICA. For reference mapping, the Lung-Kim dataset is publicly accessible via the Curated Cancer Cell Atlas (https://www.weizmann.ac.il/sites/3CA/lung). The processed COVID-19 dataset was accessed at https://github.com/theislab/scarches-reproducibility. For the perturbation prediction task, the Norman and Adamson datasets were retrieved from the following links: https://dataverse.harvard.edu/api/access/datafile/6154020 and https://dataverse.harvard.edu/api/access/datafile/6154417. The Replogle dataset was retrieved from https://gwps.wi.mit.edu/. For the batch integration task, the PBMC 10k dataset was retrieved from the scVI tools (https://scvi-tools.org/) using the API scvi.data.pbmc_dataset. The perirhinal cortex dataset was retrieved from the CELLxGENE Human Brain Cell Atlas version 1.0 (https://cellxgene.cziscience.com/collections/283d65eb-dd53-496d-adb7-7570c7caa443). For the multi-omic integration task, the 10x Multiome PBMC dataset was retrieved from https://scglue.readthedocs.io/en/latest/data.html. The BMMC dataset is accessible from the GEO database via accession number GSE194122. The ASAP PBMC dataset was retrieved from https://github.com/PeterZZQ/scMoMaT/tree/main/data/real/ASAP-PBMC. For GRN analysis, the processed Immune Human dataset was accessed from https://doi.org/10.6084/m9.figshare.12420968.v8. All processed datasets can be accessed at https://github.com/bowang-lab/scGPT and https://doi.org/10.6084/m9.figshare.24954519.v1 (ref. 73).
Code availability
The codebase for scGPT is publicly available at https://github.com/bowang-lab/scGPT and at the Zenodo repository74 (https://doi.org/10.5281/zenodo.10466117) with the MIT License.
References
Silverman, A. D., Karim, A. S. & Jewett, M. C. Cell-free gene expression: an expanded repertoire of applications. Nat. Rev. Genet. 21, 151–170 (2020).
Preissl, S., Gaulton, K. J. & Ren, B. Characterizing cis-regulatory elements using single-cell epigenomics. Nat. Rev. Genet. 24, 21–43 (2022).
Ding, J., Sharon, N. & Bar-Joseph, Z. Temporal modelling using single-cell transcriptomics. Nat. Rev. Genet. 23, 355–368 (2022).
Wagner, D. E. & Klein, A. M. Lineage tracing meets single-cell omics: opportunities and challenges. Nat. Rev. Genet. 21, 410–427 (2020).
Regev, A. Science Forum: the Human Cell Atlas. eLife 6, e27041 (2017).
Han, X. Mapping the mouse cell atlas by Microwell-seq. Cell 172, 1091–1107 (2018).
Angerer, P. et al. Single cells make big data: new challenges and opportunities in transcriptomics. Curr. Opin. Syst. Biol. 4, 85–91 (2017).
Subramanian, I., Verma, S., Kumar, S., Jere, A. & Anamika, K. Multi-omics data integration, interpretation, and its application. Bioinform. Biol. Insights 14, 1177932219899051 (2020).
Miao, Z., Humphreys, B. D., McMahon, A. P. & Kim, J. Multi-omics integration in the age of million single-cell data. Nat. Rev. Nephrol. 17, 710–724 (2021).
Lotfollahi, M., Wolf, F. A. & Theis, F. J. scGen predicts single-cell perturbation responses. Nat. Methods 16, 715–721 (2019).
Lotfollahi, M. Predicting cellular responses to complex perturbations in high-throughput screens. Mol. Syst. Biol. 19, e11517 (2023).
Lotfollahi, M. Mapping single-cell data to reference atlases by transfer learning. Nat. Biotechnol. 40, 121–130 (2022).
Cao, Z.-J. & Gao, G. Multi-omics single-cell data integration and regulatory inference with graph-linked embedding. Nat. Biotechnol. 40, 1458–1466 (2022).
Zhang, Z. et al. scMoMat jointly performs single cell mosaic integration and multi-modal bio-marker detection. Nat. Commun. 14, 384 (2023).
Bommasani, R. et al. On the opportunities and risks of foundation models. Preprint at https://doi.org/10.48550/arXiv.2108.07258 (2021).
Moor, M. et al. Foundation models for generalist medical artificial intelligence. Nature 616, 259–265 (2023).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 6000–6010 (NeurIPS, 2017).
Ramesh, A., Dhariwal, P., Nichol, A., Chu, C. & Chen, M. Hierarchical text-conditional image generation with CLIP latents. Preprint at https://doi.org/10.48550/arXiv.2204.06125 (2022).
Brown, T. Language models are few-shot learners. Adv. Neural. Inf. Process. Syst. 1877–1901 (NeurIPS, 2020).
OpenAI team. GPT-4 technical report. Preprint at https://doi.org/10.48550/arXiv.2303.08774 (2023).
Avsec, Ž. et al. Effective gene expression prediction from sequence by integrating long-range interactions. Nat. Methods 18, 1196–1203 (2021).
Gururangan, S. et al. Don’t stop pretraining: adapt language models to domains and tasks. In Proc. 58th Annual Meeting of the Association for Computational Linguistics 8342–8360 (ACL, 2020).
Qiu, X. et al. Pre-trained models for natural language processing: a survey. Sci. China Technol. Sci. 63, 1872–1897 (2020).
Liu, J., Fan, Z., Zhao, W. & Zhou, X. Machine intelligence in single-cell data analysis: advances and new challenges. Front. Genet. 12, 655536 (2021).
Oller-Moreno, S., Kloiber, K., Machart, P. & Bonn, S. Algorithmic advances in machine learning for single-cell expression analysis. Curr. Opin. Syst. Biol. 25, 27–33 (2021).
Ji, Y., Lotfollahi, M., Wolf, F. A. & Theis, F. J. Machine learning for perturbational single-cell omics. Cell Syst. 12, 522–537 (2021).
Theodoris, C. V. et al. Transfer learning enables predictions in network biology. Nature 618, 616–624 (2023).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://doi.org/10.48550/arXiv.1802.03426 (2018).
Schirmer, L. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature 573, 75–82 (2019).
Cheng, S. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 184, 792–809 (2021).
Chen, J. et al. Transformer for one stop interpretable cell type annotation. Nat. Commun. 14, 223 (2023).
Yang, F. et al. scBERT as a large-scale pretrained deep language model for cell type annotation of single-cell RNA-seq data. Nat. Mach. Intell. 4, 852–866 (2022).
Adamson, B. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882 (2016).
Replogle, J. M. Mapping information-rich genotype–phenotype landscapes with genome-scale Perturb-seq. Cell 185, 2559–2575 (2022).
Norman, T. M. et al. Exploring genetic interaction manifolds constructed from rich single-cell phenotypes. Science 365, 786–793 (2019).
Roohani, Y., Huang, K. & Leskovec, J. Predicting transcriptional outcomes of novel multigene perturbations with GEARS. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01905-6 (2023).
Traag, V. A., Waltman, L. & Van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Gayoso, A. A Python library for probabilistic analysis of single-cell omics data. Nat. Biotechnol. 40, 163–166 (2022).
Siletti, K. Transcriptomic diversity of cell types across the adult human brain. Science 382, eadd7046 (2023).
PBMC from a healthy donor, single cell multiome ATAC gene expression demonstration data by Cell Ranger ARC 1.0.0. 10X Genomics https://support.10xgenomics.com/single-cell-multiome-atac-gex/datasets/1.0.0/pbmc_granulocyte_sorted_10k (2020).
Hao, Y. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Luecken, M. et al. A sandbox for prediction and integration of DNA, RNA, and proteins in single cells. In Proceedings of the Neural Information Processing Systems Track on Datasets and Benchmarks 13 (NeurIPS, 2021).
Mimitou, E. P. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol. 39, 1246–1258 (2021).
Pratapa, A., Jalihal, A. P., Law, J. N., Bharadwaj, A. & Murali, T. M. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat. Methods 17, 147–154 (2020).
Choo, S. Y. The HLA system: genetics, immunology, clinical testing, and clinical implications. Yonsei Med. J. 48, 11–23 (2007).
Norman, P. S. Immunobiology: the immune system in health and disease. J. Allergy Clin. Immunol. 96, 274 (1995).
Luecken, M. D. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods 19, 41–50 (2022).
Zou, Z., Ohta, T., Miura, F. & Oki, S. ChIP-Atlas 2021 update: a data-mining suite for exploring epigenomic landscapes by fully integrating ChIP–seq, ATAC-seq and Bisulfite-seq data. Nucleic Acids Res. 50, W175–W182 (2022).
Yang, H., Niemeijer, M., van de Water, B. & Beltman, J. B. ATF6 is a critical determinant of CHOP dynamics during the unfolded protein response. iScience 23, 100860 (2020).
Yoshida, H. et al. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20, 6755–6767 (2000).
Kaplan, J. et al. Scaling laws for neural language models. Preprint at https://doi.org/10.48550/arXiv.2001.08361 (2020).
Sarkar, A. & Stephens, M. Separating measurement and expression models clarifies confusion in single-cell RNA sequencing analysis. Nat. Genet. 53, 770–777 (2021).
Haque, A., Engel, J., Teichmann, S. A. & Lönnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 9, 1–12 (2017).
Devlin, J., Chang, M. W., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In Proc. 2019 Conference of the North American Chapter of the Association for Computational Linguistics 4171–4186 (ACL, 2019).
Dao, T., Fu, D., Ermon, S., Rudra, A. & Ré, C. FlashAttention: fast and memory-efficient exact attention with IO-Awareness. Adv. Neural. Inf. Process. Syst. 16344–16359 (NeurIPS, 2022).
Wang, S., Li, B. Z., Khabsa, M., Fang, H. & Ma, H. Linformer: self-attention with linear complexity. Preprint at https://doi.org/10.48550/arXiv.2006.04768 (2020).
Katharopoulos, A., Vyas, A., Pappas, N. & Fleuret, F. Transformers are RNNs: fast autoregressive transformers with linear attention. In Proc. 37th International Conference on Machine Learning 5156–5165 (PMLR, 2020).
Liu, Y. RoBERTa: a robustly optimized BERT pretraining approach. Preprint at https://doi.org/10.48550/arXiv.1907.11692 (2019).
Bubeck, S. et al. Sparks of artificial general intelligence: early experiments with GPT-4. Preprint at https://doi.org/10.48550/arXiv.2303.12712 (2023).
Liu, C. et al. Guided similarity separation for image retrieval. Adv. Neural. Inf. Process. Syst. 1556–1566 (NeurIPS, 2019).
Eisenstein, M. Single-cell RNA-seq analysis software providers scramble to offer solutions. Nat. Biotechnol. 38, 254–257 (2020).
Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020).
Ganin, Y. & Lempitsky, V. Unsupervised domain adaptation by backpropagation. In Proc. 32nd International Conference on Machine Learning 1180–1189 (PMLR, 2015).
Ceglia, N. Identification of transcriptional programs using dense vector representations defined by mutual information with GeneVector. Nat. Commun. 14, 4400 (2023).
Kim, N. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 11, 2285 (2020).
Paszke, A. PyTorch: an imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Sys. 1–12 (NeurIPS, 2019).
Wolf, F. A., Angerer, P. & Theis, F. J. Scanpy: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Danese, A. et al. EpiScanpy: integrated single-cell epigenomic analysis. Nat. Commun. 12, 5228 (2021).
Fang, Z., Liu, X. & Peltz, G. GSEApy: a comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics 39, btac757 (2023).
Wang, C. Processed datasets used in the scGPT foundation model. Figshare https://doi.org/10.6084/m9.figshare.24954519.v1 (2024).
Cui, H., Wang, C. & Pang, K. Codebase for scGPT: towards building a foundation model for single-cell multi-omics using generative AI. Zenodo https://doi.org/10.5281/zenodo.10466117 (2024).
Acknowledgements
We appreciate valuable feedback from L. Zhang during the writing of the manuscript. The UMAP illustrations in Fig. 1a were created using CELLxGENE Annotate (https://github.com/chanzuckerberg/cellxgene). Fig. 1d was created with BioRender (https://www.biorender.com). This work was supported by funding from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2020-06189 and DGECR-2020-00294, B.W.), the CIFAR AI Chairs Program (B.W.) and the Peter Munk Cardiac Centre AI Fund at the University Health Network (B.W.). This research was undertaken, in part, thanks to funding from the Canada Research Chairs Program. H.M. is supported by a doctoral fellowship from the Natural Sciences and Engineering Research Council of Canada.
Author information
Authors and Affiliations
Contributions
H.C. developed the concept of the work and contributed to design and implementation of the algorithm. C.W. and K.P. contributed to design and implementation of the algorithm. H.C., C.W., H.M., K.P. and F.L. contributed to the analysis of computational experiments. H.C. and C.W. drafted the initial version of the manuscript. H.C., C.W., H.M., K.P., F.L. and B.W. contributed to revision of the work. N.D. contributed to design of the algorithm. B.W. contributed to the conception and design of the work.
Corresponding author
Ethics declarations
Competing interests
B.W. is on the advisory board of Vevo Therapeutics. N.D. is an employee of Microsoft and holds equity in the company. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Lin Tang, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–12, Tables 1–7 and Figs. 1–13
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cui, H., Wang, C., Maan, H. et al. scGPT: toward building a foundation model for single-cell multi-omics using generative AI. Nat Methods (2024). https://doi.org/10.1038/s41592-024-02201-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41592-024-02201-0