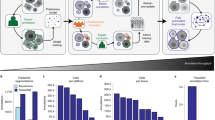
Abstract
Spatial transcriptomics promises to greatly improve our understanding of tissue organization and cell–cell interactions. While most current platforms for spatial transcriptomics only offer multi-cellular resolution, with 10–15 cells per spot, recent technologies provide a much denser spot placement leading to subcellular resolution. A key challenge for these newer methods is cell segmentation and the assignment of spots to cells. Traditional image-based segmentation methods are limited and do not make full use of the information profiled by spatial transcriptomics. Here we present subcellular spatial transcriptomics cell segmentation (SCS), which combines imaging data with sequencing data to improve cell segmentation accuracy. SCS assigns spots to cells by adaptively learning the position of each spot relative to the center of its cell using a transformer neural network. SCS was tested on two new subcellular spatial transcriptomics technologies and outperformed traditional image-based segmentation methods. SCS achieved better accuracy, identified more cells and provided more realistic cell size estimation. Subcellular analysis of RNAs using SCS spot assignments provides information on RNA localization and further supports the segmentation results.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data used in this study have been previously published. The spatial transcriptomics data and nucleus staining images for the Stereo-seq dataset are available in the MOSTA data portal (https://db.cngb.org/stomics/mosta/download/) with file names Mouse_brain_Adult_GEM_bin1.tsv.gz and Mouse_brain_Adult.tif. The spatial transcriptomics data for the Seq-scope dataset are available in the Gene Expression Omnibus database under accession number GSE169706. Tiles 2104, 2105, 2106 and 2107 were used in this study. The H&E staining images for the Seq-scope dataset are available at Deep Blue Data (https://doi.org/10.7302/cjfe-wa35). The seqFISH+ NIH/3T3 cell line data are available at Zenodo (https://doi.org/10.5281/zenodo.2669683). The spatial transcriptomics data of the MERFISH human brain dataset are available at Dryad (https://doi.org/10.5061/dryad.x3ffbg7mw). The section of H22.26.401.MTG.4000 was used in this study. Experimental RNA subcellular localization data are available in the RNALocate v.2.0 database (https://www.rna-society.org/rnalocate/download.html).
Code availability
The source code of SCS is publicly available at https://github.com/chenhcs/SCS. The following open source Python (v.3.9.7) packages were used to build SCS: anndata (v.0.7.5), matplotlib (v.3.5.0), numpy (v.1.22.4), pandas (v.1.3.4), scanpy (v.1.8.2), scikit-learn (v.1.0.1), scipy (v.1.7.2) and tensorflow (v.2.8.2). The open source software Spateo (v.0.0.0) was used to align staining image pixels with spatial transcriptomics spots. Watershed implemented in Spateo (v.0.0.0), open source Python packages DeepCell (v.0.12.3), Cellpose (v.2.1.1), StarDist (0.8.3), Baysor (v.0.5.2) and JSTA (v.0.0.0) were applied to segment staining images and compared with SCS.
References
Li, D., Ding, J. & Bar-Joseph, Z. Identifying signaling genes in spatial single-cell expression data. Bioinformatics 37, 968–975 (2021).
Teng, H., Yuan, Y. & Bar-Joseph, Z. Clustering spatial transcriptomics data. Bioinformatics 38, 997–1004 (2022).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792 (2022).
Cho, Chun-Seok et al. Microscopic examination of spatial transcriptome using Seq-scope. Cell 184, 3559–3572 (2021).
Dayao, M. T., Brusko, M., Wasserfall, C. & Bar-Joseph, Z. Membrane marker selection for segmenting single cell spatial proteomics data. Nat. Commun. 13, 1999 (2022).
Petukhov, V. et al. Cell segmentation in imaging-based spatial transcriptomics. Nat. Biotechnol. 40, 345–354 (2022).
Littman, R. et al. Joint cell segmentation and cell type annotation for spatial transcriptomics. Mol. Syst. Biol. 17, e10108 (2021).
Beucher, S. Use of watersheds in contour detection. In Proc. International Workshop on Image Processing 17–21 (CCETT, 1979).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Bannon, D. et al. Deepcell kiosk: scaling deep learning–enabled cellular image analysis with kubernetes. Nat. Methods 18, 43–45 (2021).
Schmidt, U., Weigert, M., Broaddus, C. & Myers, G. Cell detection with star-convex polygons. In Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Proc. International Conference on Medical Image Computing and Computer-Assisted Intervention 265–273 (Springer, 2018).
Greenwald, N. F. et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 40, 555–565 (2022).
Gilman, J. P., Medalla, M. & Luebke, J. I. Area-specific features of pyramidal neurons-a comparative study in mouse and rhesus monkey. Cerebral Cortex 27, 2078–2094 (2017).
McInnes, L., Healy, J., Saul, N. & Großberger, L. UMAP: uniform manifold approximation and projection. J. Open Source Softw. 3, 861 (2018).
Junatas, K. L. et al. Stereological analysis of size and density of hepatocytes in the porcine liver. J. Anatomy 230, 575–588 (2017).
Eng, Chee-HuatLinus et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqfish+. Nature 568, 235–239 (2019).
Fang, R. et al. Conservation and divergence of cortical cell organization in human and mouse revealed by merfish. Science 377, 56–62 (2022).
Cable, D. M. et al. Cell type-specific inference of differential expression in spatial transcriptomics. Nat. Methods 19, 1076–1087 (2022).
Yuan, Y. & Bar-Joseph, Z. Gcng: graph convolutional networks for inferring gene interaction from spatial transcriptomics data. Genome Biol. 21, 300 (2020).
Qin, Y. et al. A multi-scale map of cell structure fusing protein images and interactions. Nature 600, 536–542 (2021).
Cui, T. et al. RNAlocate v2. 0: an updated resource for RNA subcellular localization with increased coverage and annotation. Nucleic Acids Res. 50, D333–D339 (2022).
Korostowski, L., Sedlak, N. & Engel, N. The kcnq1ot1 long non-coding RNA affects chromatin conformation and expression of kcnq1, but does not regulate its imprinting in the developing heart. PLOS Genetics 8, e1002956 (2012).
Clemson, C. M. et al. An architectural role for a nuclear noncoding RNA: Neat1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726 (2009).
Van Weering, JanR. T., Toonen, R. F. & Verhage, M. The role of rab3a in secretory vesicle docking requires association/dissociation of guanidine phosphates and munc18-1. PLoS ONE 2, e616 (2007).
Chanaday, N. L. & Kavalali, E. T. Synaptobrevin-2 dependent regulation of single synaptic vesicle endocytosis. Mol. Biol. Cell 32, 1818–1823 (2021).
Engel, K. L., Arora, A., Goering, R., Lo, Hei-YongG. & Taliaferro, J. M. Mechanisms and consequences of subcellular RNA localization across diverse cell types. Traffic 21, 404–418 (2020).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792 (2022).
Cho, Chun-Seok et al. Microscopic examination of spatial transcriptome using Seq-scope. Cell 184, 3559–3572 (2021).
Vaswani, A. et al. Attention is all you need. Adv. Neural Info. Proc. Syst. 30, 5998–6008 (2017).
Hendrycks, D. & Gimpel, K. Gaussian error linear units (gelus). Preprint at arXiv1606.08415 (2016).
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I. & Salakhutdinov, R. Dropout: a simple way to prevent neural networks from overfitting. J. Machine Learn. Res. 15, 1929–1958 (2014).
Ba, J. L., Kiros, J. R. & Hinton, G. E. Layer normalization. Preprint at arXiv1607.06450 (2016).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 770–778 (IEEE, 2016).
Li, G. et al. 3D cell nuclei segmentation based on gradient flow tracking. BMC Cell Biol. 8, 40 (2007).
Cui, T. et al. RNAlocate v2. 0: an updated resource for RNA subcellular localization with increased coverage and annotation. Nucleic Acids Res. 50, D333–D339 (2022).
Wolf, F. A., Angerer, P. & Theis, F. J. Scanpy: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Acknowledgements
We thank L. Liu, J.H. Lee and R. Fang for sharing the Stereo-seq, Seq-scope and MERFISH data, respectively, and for advising us on how to process these datasets. This work was partially supported by National Institutes of Health grant nos. OT2OD026682, 1U54AG075931 and 1U24CA268108 to Z.B.-J.
Author information
Authors and Affiliations
Contributions
H.C., D.L. and Z.B.-J. conceptualized and designed the study. H.C., D.L. and Z.B.-J. designed the algorithm and methodology. H.C. developed the software of SCS with supervision from Z.B.-J. H.C. and D.L. performed evaluations and result analyses. H.C., D.L. and Z.B.-J. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Qinghua Jiang, Jonas Maaskola and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Rita Strack, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Visualization of barcoded spots (red dots) overlaid on the staining image for one patch of the Stereo-seq dataset.
The darker the color of each dot, the higher the total number of RNA molecules captured in the spot. Color scale for the RNA counts is shown in the color bar. Spot placement achieves sub-cellular resolution. The boundaries of cells cannot be visually identified from spots. The visualization on all the 87 patches of the Stereo-seq dataset show similar patterns. Scale bar: 100 μm.
Extended Data Fig. 2 Distribution of the total UMI counts of barcoded spots on the Stereo-seq dataset.
This dataset achieves an average of 3.3 unique molecular identifier (UMI) counts per spot.
Extended Data Fig. 3 Visualization of barcoded spots (red dots) overlaid on the staining image for one section of the Seq-scope dataset.
The darker the color of each dot, the higher the total number of RNA molecules captured in the spot. Color scale for the RNA counts is shown in the color bar. Spot placement achieves sub-cellular resolution. The boundaries of cells cannot be visually identified from spots. The visualization on all the four sections of the Seq-scope dataset show similar patterns. Scale bar: 100 μm.
Extended Data Fig. 4 Distribution of the total UMI counts of barcoded spots on the Seq-scope dataset.
This dataset achieves an average of 5.7 unique molecular identifier (UMI) counts per spot.
Extended Data Fig. 5 Experimental evidence for differentially localized RNAs identified by SCS.
a, Agreement between the experimental localization evidence and SCS identification of genes whose RNAs are differentially localized for Stereo-seq. b, Agreement between the experimental evidence and SCS identifications for the Seq-scope dataset.
Supplementary information
Supplementary Information
Supplementary Notes, Figs. 1–12 and Table 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, H., Li, D. & Bar-Joseph, Z. SCS: cell segmentation for high-resolution spatial transcriptomics. Nat Methods 20, 1237–1243 (2023). https://doi.org/10.1038/s41592-023-01939-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-023-01939-3
This article is cited by
-
BIDCell: Biologically-informed self-supervised learning for segmentation of subcellular spatial transcriptomics data
Nature Communications (2024)