Abstract
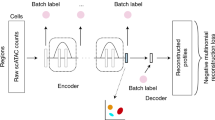
Single-cell assay for transposase-accessible chromatin using sequencing (scATAC) shows great promise for studying cellular heterogeneity in epigenetic landscapes, but there remain important challenges in the analysis of scATAC data due to the inherent high dimensionality and sparsity. Here we introduce scBasset, a sequence-based convolutional neural network method to model scATAC data. We show that by leveraging the DNA sequence information underlying accessibility peaks and the expressiveness of a neural network model, scBasset achieves state-of-the-art performance across a variety of tasks on scATAC and single-cell multiome datasets, including cell clustering, scATAC profile denoising, data integration across assays and transcription factor activity inference.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
We used only public datasets in this study. We downloaded the processed peak set for Buenrostro2018 generated by Chen et al. at https://github.com/pinellolab/scATAC-benchmarking/blob/master/Real_Data/Buenrostro_2018/input/combined.sorted.merged.bed. We downloaded the aligned bam files from https://github.com/pinellolab/scATAC-benchmarking/tree/master/Real_Data/Buenrostro_2018/input/sc-bams_nodup. The original datasets are from the Gene Expression Omnibus (GEO) under accession code GSE96769. We downloaded the 10x multiome datasets from 10x Genomics at https://support.10xgenomics.com/single-cell-multiome-atac-gex/datasets/2.0.0/pbmc_granulocyte_sorted_3k for the PBMC dataset and https://support.10xgenomics.com/single-cell-multiome-atac-gex/datasets/2.0.0/e18_mouse_brain_fresh_5k for the mouse brain dataset. We downloaded the processed peak-by-cell matrix from sci-ATAC human atlas (GEO accession code GSE184461) stored at http://renlab.sdsc.edu/kai/Key_Processed_Data/Cell_by_cCRE/. spear-ATAC preprocessed count matrix ‘K562-Pilot-scATAC-Peak-Matrix-SE.rds’ was downloaded from GEO (accession code GSE168851). Source data are provided with this paper.
Code availability
Code for training and using the scBasset model can be found at https://github.com/calico/scBasset. Instructions and tutorials are provided at the GitHub repository for how to train scBasset models from anndata and to compute cell embeddings, denoise accessibility profiles, perform TF activity inference and ISM from a trained scBasset model. A trained scBasset model for the Buenrostro2018 dataset is available in the kipoi model zoo (https://github.com/kipoi/models/tree/master/scbasset).
Change history
23 December 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41592-022-01754-2
References
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Satpathy, A. T. et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925–936 (2019).
Miao, Z. et al. Single cell regulatory landscape of the mouse kidney highlights cellular differentiation programs and renal disease targets. Nat. Commun. 12, 2277 (2021).
Cusanovich, D. A. et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–1324 (2018).
Bravo González-Blas, C. et al. cisTopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nat. Methods 16, 397–400 (2019).
Chen, H. et al. Assessment of computational methods for the analysis of single-cell ATAC-seq data. Genome Biol. 20, 241 (2019).
Pliner, H. A. et al. Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol. Cell 71, 858–871 (2018).
Xiong, L. et al. SCALE method for single-cell ATAC-seq analysis via latent feature extraction. Nat. Commun. 10, 4576 (2019).
Schep, A. N., Wu, B., Buenrostro, J. D. & Greenleaf, W. J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017).
de Boer, C. G. & Regev, A. BROCKMAN: deciphering variance in epigenomic regulators by k-mer factorization. BMC Bioinf. 19, 253 (2018).
Kelley, D. R., Snoek, J. & Rinn, J. L. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 26, 990–999 (2016).
Zhou, J. & Troyanskaya, O. G. Predicting effects of noncoding variants with deep-learning-based sequence model. Nat. Methods 12, 931–934 (2015).
Kelley, D. R. et al. Sequential regulatory activity prediction across chromosomes with convolutional neural networks. Genome Res. 28, 739–750 (2018).
Avsec, Ž. et al. Base-resolution models of transcription-factor binding reveal soft motif syntax. Nat. Genet. 53, 354–366 (2021).
Buenrostro, J. D. et al. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 173, 1535–1548.e16 (2018).
Qin, Q. et al. Lisa: inferring transcriptional regulators through integrative modeling of public chromatin accessibility and ChIP-seq data. Genome Biol. 21, 32 (2020).
Wu, K. E., Yost, K. E., Chang, H. Y. & Zou, J. BABEL enables cross-modality translation between multiomic profiles at single-cell resolution. Proc. Natl Acad. Sci. USA 118, e2023070118 (2021).
Lotfollahi, M. et al. Mapping single-cell data to reference atlases by transfer learning. Nat. Biotechnol. 40, 121–130 (2022).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Büttner, M., Miao, Z., Wolf, F. A., Teichmann, S. A. & Theis, F. J. A test metric for assessing single-cell RNA-seq batch correction. Nat. Methods 16, 43–49 (2019).
Luecken, M. D. et al. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods 19, 41–50 (2022).
Li, Z. et al. Chromatin-accessibility estimation from single-cell ATAC-seq data with scOpen. Nat. Commun. 12, 6386 (2021).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Kiss, M. et al. Retinoid X receptor suppresses a metastasis-promoting transcriptional program in myeloid cells via a ligand-insensitive mechanism. Proc. Natl Acad. Sci. USA 114, 10725–10730 (2017).
Pierce, S. E., Granja, J. M. & Greenleaf, W. J. High-throughput single-cell chromatin accessibility CRISPR screens enable unbiased identification of regulatory networks in cancer. Nat. Commun. 12, 2969 (2021).
Tuan, D., Solomon, W., Li, Q. & London, I. M. The ‘β-like-globin’ gene domain in human erythroid cells. Proc. Natl Acad. Sci. USA 82, 6384–6388 (1985).
Li, Q., Peterson, K. R., Fang, X. & Stamatoyannopoulos, G. Locus control regions. Blood 100, 3077–3086 (2002).
Tallack, M. R. et al. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 20, 1052–1063 (2010).
Zhang, K. et al. A single-cell atlas of chromatin accessibility in the human genome. Cell 184, 5985–6001 (2021).
Kelley, D. R. Cross-species regulatory sequence activity prediction. PLoS Comput. Biol. 16, e1008050 (2020).
Janssens, J. et al. Decoding gene regulation in the fly brain. Nature 601, 630–636 (2022).
Shrikumar, A. et al. Technical note on transcription factor motif discovery from importance scores (TF-MoDISco) version 0.5.6.5. arXiv. https://arxiv.org/abs/1811.00416 (2018).
Lal, A. et al. Deep learning-based enhancement of epigenomics data with AtacWorks. Nat. Commun. 12, 1507 (2021).
Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
Hou, W., Ji, Z., Ji, H. & Hicks, S. C. A systematic evaluation of single-cell RNA-sequencing imputation methods. Genome Biol. 21, 218 (2020).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Fang, R. et al. Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun. https://doi.org/10.1038/s41467-021-21583-9 (2021).
Ashuach, T., Reidenbach, D. A., Gayoso, A. & Yosef, N. PeakVI: A deep generative model for single-cell chromatin accessibility analysis. Cell Rep. Methods 2, 100182 (2022).
Liu, Q., Chen, S., Jiang, R. & Wong, W. H. Simultaneous deep generative modelling and clustering of single-cell genomic data. Nat. Mach. Intell. https://doi.org/10.1038/s42256-021-00333-y (2021).
van Dijk, D. et al. Recovering gene interactions from single-cell data using data diffusion. Cell https://doi.org/10.1016/j.cell.2018.05.061 (2018).
Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020).
Jiang, M., Anderson, J., Gillespie, J. & Mayne, M. uShuffle: a useful tool for shuffling biological sequences while preserving the k-let counts. BMC Bioinf. 9, 192 (2008).
Weirauch, M. T. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014).
Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Acknowledgements
We thank V. Agarwal, J. Kimmel and M. Mohamed for feedback on the manuscript. We thank S. Spock for feedback on the code. We also thank N. Bernstein and A. Odak for helpful discussions.
Author information
Authors and Affiliations
Contributions
D.R.K. conceived the project. H.Y. and D.R.K. developed the model. H.Y. performed the analysis. H.Y. and D.R.K prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
H.Y. and D.R.K. are paid employees of Calico Life Sciences.
Peer review
Peer review information
Nature Methods thanks Luca Pinello, Qiangfeng Cliff Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling editor: Lin Tang, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Buenrostro2018 cell embeddings.
t-SNE visualization of different cell embedding methods on Buenrostro2018, including: chromVAR motif, chromVAR kmer (k = 6), PCA, cicero (LSI), ArchR, snapATAC, cisTopic, scDEC, SCALE, peakVI and scBasset.
Extended Data Fig. 2 10x multiome PBMC cell embeddings.
UMAP visualization of different cell embedding methods on the 10x multiome PBMC dataset, including: chromVAR_motif, chromVAR_kmer (k = 6), PCA, cicero (LSI), ArchR, snapATAC, cisTopic, scDEC, SCALE, peakVI and scBasset.
Extended Data Fig. 3 scBasset batch correction on chemistry-mixed PBMC.
a) Model architecture of scBasset-BC. b) Performance of scBasset batch correction models trained on the chemistry-mixed PBMC data. We trained scBasset-BC models with increasing L2 penalty (from 0 to 1e-2). Batch-mixing is measured with kBET and iLISI, and conservation of biological variation is measured with label score. c) Performance comparison of different batch correction methods on the chemistry-mixed PBMC data. Harmony is applied on either PCA, Harmony(PCA), or scBasset embeddings, Harmony(scBasset). Performance is evaluated by kBET, iLISI and label score. d) UMAPs of scBasset batch correction with different L2 penalties on the chemistry-mixed PBMC data. We selected L2 = 1e-6 as the fin42al scBasset-BC model. e) UMAPs of different batch correction methods on the chemistry-mixed PBMC data.
Extended Data Fig. 4 scBasset batch correction on Buenrostro2018.
a) scBasset batch correction performance as a function of L2 penalty on Buenrostro2018 dataset. Performance is evaluated by kBET, iLISI and label score. b) Performance comparison of different batch correction methods on Buenrostro2018 data. Harmony is applied on either PCA, Harmony(PCA), or scBasset embeddings, Harmony(scBasset). Performance is evaluated by kBET, iLISI and label score. c) UMAPs of scBasset batch correction with different L2 penalties on Buenrostro2018 data. We selected L2 = 1e-8 as the final scBasset-BC model. d) UMAPs of different batch correction methods on Buenrostro2018 data.
Extended Data Fig. 5 Additional scBasset denoising and integration results.
a) Comparison of different denoising methods in Buenrostro2018 dataset as evaluated by label score and cell type average Silhouette width (cell type ASW). b) Correlation between gene accessibility score and gene expression across genes for each cell before (x-axis) and after scBasset denoising (y-axis) for the multiome mouse brain dataset. Cells are colored by sequencing depth. c) Left, Comparison of different denoising methods in multiome mouse brain dataset as evaluated by label score and cell type ASW. Right, comparison of different denoising methods in multiome mouse brain dataset as evaluated by correlation between scVI-denoised RNA and denoised ATAC profiles across genes per cell (correlation per cell (RNA,ATAC)), and correlation between scVI-denoised RNA and denoised ATAC profiles across cells per gene (correlation per gene (RNA,ATAC)). d) Integration performance comparison in multiome 10x PBMC dataset when (i) both RNA and ATAC profiles are raw; (ii) only RNA profile is denoised with scVI; (iii) only ATAC profile is denoised with scBasset; and (iv) both RNA and ATAC profiles are denoised. n = 2714 cells for each boxplot. The boxplot shows min and max as whiskers (excluding outliers), 1st and 3rd quartiles as boxes and median in the center. Outliers (> 1.5x interquartile range away from the box) are not shown. e) UMAPs of RNA and ATAC co-embedding after integration for 10x multiome mouse brain dataset. Left, integration performed on RNA (blue) and raw ATAC (red) profile embeddings. Right, integration performed on RNA (blue) and scBasset-denoised ATAC (red).
Extended Data Fig. 6 Motif activity inference using scBasset and chromVAR on Buenrostro2018 dataset.
a) UMAPs showing scBasset-predicted TF activity. b) Boxplots showing scBasset-predicted TF activity by cell type. c) UMAPs showing chromVAR-predicted TF activity. d) Boxplots showing chromVAR-predicted TF activity per cell type. For boxplots in B) and D), n = 502, 402, 344, 160, 142, 141, 138, 78, 64, 60 cells for each of CMP, GMP, HSC, LMPP, MPP, pDC, MEP, CLP, mono, UNK cell types. The box extends from the first quartile (Q1) to the third quartile (Q3) of the data, with a line at the median. The whiskers extend from the box by 1.5x the interquartile range (IQR). Outliers are not shown.
Extended Data Fig. 7 TF expression and activity correlation for the 10x multiome datasets.
Scatterplots of correlations between chromVAR-inferred activity and expression (x-axis) versus correlations of scBasset-inferred TF activity and expression (y-axis) for activating TFs (left) and repressive TFs (right) in the 10x multiome PBMC (top) and 10x multiome mouse brain (bottom). Activating TFs are TFs for which both scBasset and chromVAR agree on a positive correlation between TF expression and activity. Repressive TFs are TFs for which both scBasset and chromVAR agree on a negative correlation between TF expression and activity. A one-sided Wilcoxon signed rank test was performed.
Extended Data Fig. 8 Motif activity inference using scBasset and chromVAR on 10x multiome PBMC dataset.
UMAP visualization of TF expression (left), scBasset TF activity (middle), and chromVAR TF activity (right) for additional known PBMC regulators. Pearson correlation between inferred TF activity and expression are shown in the titles.
Extended Data Fig. 9 Comparison of scBasset and chromVAR in TF perturbation experiments.
a) Embeddings of K562 cells transfected by a pool of 9 CRISPRi sgRNAs targeting GATA1 (sgGATA1) and GATA2 (sgGATA2) and 9 inert sgRNA controls (sgNT). b) Performance comparison of scBasset and chromVAR in distinguishing sgGATA1 cells from sgNT cells in ROC curves. c) Performance comparison of scBasset and chromVAR in distinguishing sgGATA2 cells from sgNT cells in ROC curves.
Extended Data Fig. 10 scBasset result on sci-ATAC human cell atlas.
a) UMAP of sci-ATAC human cell atlas. Cells colored by tissue of origin. b-d) Runtime, peak CPU memory and GPU memory usage of scBasset as a function of the number of cells in the dataset.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary Notes and Supplementary Figs. 1–9
Source data
Source Data Fig. 1
Statistical Source Data.
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Statistical Source Data.
Source Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 3
Statistical Source Data.
Source Data Extended Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 6
Statistical Source Data.
Source Data Extended Data Fig. 7
Statistical Source Data.
Source Data Extended Data Fig. 10
Statistical Source Data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yuan, H., Kelley, D.R. scBasset: sequence-based modeling of single-cell ATAC-seq using convolutional neural networks. Nat Methods 19, 1088–1096 (2022). https://doi.org/10.1038/s41592-022-01562-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-022-01562-8
This article is cited by
-
Cell-type-directed design of synthetic enhancers
Nature (2024)
-
Single-cell multi-ome regression models identify functional and disease-associated enhancers and enable chromatin potential analysis
Nature Genetics (2024)
-
Genotype imputation methods for whole and complex genomic regions utilizing deep learning technology
Journal of Human Genetics (2024)
-
A fast, scalable and versatile tool for analysis of single-cell omics data
Nature Methods (2024)
-
Challenges and best practices in omics benchmarking
Nature Reviews Genetics (2024)