Abstract
The recent development of solvent- and polymer-based brain-clearing techniques has advanced our ability to visualize the mammalian nervous system in three dimensions. However, it remains challenging to image the mammalian body en bloc. Here we developed HYBRiD (hydrogel-based reinforcement of three-dimensional imaging solvent-cleared organs (DISCO)), by recombining components of organic- and polymer-based clearing pipelines. We achieved high transparency and protein retention, as well as compatibility with direct fluorescent imaging and immunostaining in cleared mammalian bodies. Using parvalbumin- and somatostatin-Cre models, we demonstrated the utility of HYBRiD for whole-body imaging of genetically encoded fluorescent reporters without antibody enhancement of signals in newborn and juvenile mice. Using K18-hACE2 transgenic mice, HYBRiD enabled perfusion-free clearing and visualization of SARS-CoV-2 infection in a whole mouse chest, revealing macroscopic and microscopic features of viral pathology in the same sample. HYBRiD offers a simple and universal solution to visualize large heterogeneous body parts or entire animals for basic and translational research.
Similar content being viewed by others
Main
Understanding physiology and disease across multiple organ systems has long been a challenge in biology. Recent developments in tissue-clearing techniques have enabled three-dimensional (3D) characterization of mammalian tissues. Primarily optimized for the brain, these methods are generally based on one of two distinct strategies. Organic solvent-based protocols (3D imaging of solvent-cleared organs (3DISCO)1, iDISCO2,3,4, uDISCO5, FDISCO6, and vDISCO7) inspired by benzyl alcohol and benzyl benzoate (BABB) and earlier methods, are rapid and robust in clearing tissues, but tend to quench fluorescence and often require additional signal amplification. On the other hand, aqueous methods, such as CUBIC8,9,10,11, and those involving polymer-reinforced scaffolds (CLARITY12,13,14,15, PACT/PARS16,17, SHIELD18,19, ACT-PRESTO20) allow better retention of endogenous signals but may require special devices or longer time to achieve complete clearing and are not suited to all tissue types (Extended Data Fig. 1a)21.
Although most of these methods clear rodent brains with ease, it remains more difficult to visualize en bloc body parts (that is, multiple organs with bone, muscle, blood vessels, and connective tissue intact in the same volume) with passive clearing as the balance of transparency and fluorescence retention is harder to obtain. Recent approaches to achieve whole-body clearing in rodents require continuous, pressurized perfusion of hazardous solvents (uDISCO5, PEGASOS22) or nanobodies (vDISCO7) over a period of several days, which can be challenging and prohibitively expensive to adopt or scale up in routine experiments.
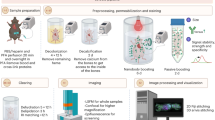
Here, we developed HYBRiD, a simple and cost-effective clearing method suitable for visualization of essentially any organ or a whole juvenile rodent by rationally recombining organic and aqueous strategies. We achieved the goal of combining hydrogel-based clearing techniques like CLARITY with solvent-based DISCO while completing whole-body clearing without perfusion of organic solvents or antibodies. HYBRiD enables clearing of large heterogeneous tissues, retention of endogenous fluorescence and protein, and minimal use of expensive, delicate, or corrosive procedures in comparison with either parental method, and it is fully compatible with direct fluorescence imaging as well as immunolabeling.
Results
Body clearing requires a different strategy than brain clearing
We performed a side-by-side comparison of several brain-clearing methods using transgenic mice that express the red fluorescent protein tdTomato under the control of parvalbumin-Cre recombinase (PV-Ai9). We chose PV-Cre as the mouse model because it expresses Cre in both the central nervous system and peripheral neurons, thus allowing us to assess tdTomato signal across the whole animal. We benchmarked CLARITY and several DISCO variants. As expected, all tested methods readily cleared the brain (Extended Data Figs. 1 and 2). We used the hindlimbs of these mice as a model to evaluate clearing in large non-brain tissues. Before clearing, we pretreated the legs with previously described decalcification (CUBIC 3.0 (ref. 10)) and decoloring (PEGASOS22) reagents. Unmodified CLARITY alone did not fully delipidate the leg after 14 days, whereas iDISCO achieved the highest transparency (Fig. 1a). However, iDISCO, together with its milder iteration FDISCO, severely bleached the tdTomato signal (Fig. 1b,c and Extended Data Fig. 1c) in brain tissue. This was mostly expected, as iDISCO was not intended to preserve fluorophores but meant for use with immunolabeling. After evaluating tdTomato fluorescence at different steps of both DISCO protocols, we found the final steps of methanol dehydration and dibenzyl ether (DBE) refractive index (RI) matching to be the primary causes of fluorescence loss in DISCO-treated samples (Fig. 1d–f), whereas replacing DBE with an aqueous RI matching solution (EasyIndex (EI) or PROTOS23) substantially rescued the tdTomato signal in FDISCO- and iDISCO-cleared samples (Fig. 1b,c and Extended Data Fig. 1c–e).
a, Representative images of whole-mount adult PV-Ai9 hindlimbs processed by the indicated protocols (six samples per condition). b, Representative confocal images of 1-mm-thick PV-Ai9 brain slices processed by the indicated protocols, that is, RI is DBE and RI is EI (six samples per condition). Native samples were fixed in PFA without clearing. Shown are tdTomato+ neurons in the neocortex. c, Quantification of fluorescence intensity of each condition in b: Native (n = 26), CLARITY (n = 31), FDISCO (EI) (n = 30), FDISCO (n = 12), iDISCO (EI) (n = 35), iDISCO (n = 15). n indicates the number of measurements from six samples per condition. Data are presented as mean raw values. Statistical significance was determined by two-tailed t-tests. a.u., arbitrary units. d–f, Representative images (d,e) and quantification of time-course analysis (f) of fluorescence quenching; 1-mm-thick brain slices were processed and stopped at individual timepoints. n = 10–16 slices from 2 animals for each step in each method. Data are presented as mean values ± s.d. MeOH, methanol. g, Schematic of the HYBRiD protocol. h, Representative images of a PV-Ai9 brain slice and hindlimb before and after FDISCO (six samples per condition). i, Representative images of FDISCO (top) or HYBRiD (bottom) brain slices (6 samples per condition) after 7 days of SDS treatment. j, Quantification of protein loss during passive SDS clearing. Data are presented as mean raw values. Statistical significance was determined by two-tailed t-tests. n = 3 samples for each group. k, Quantification of the transparency of hindlimbs. All values are mean ± s.e.m.; n = 6 samples for each group. Statistical significance (false discovery rate) between HYBRiD and FDISCO was determined by multiple unpaired t-tests with two-stage step-up. l, Representative images of a PV-Ai9 brain slice and hindlimb processed by HYBRiD from three samples. Grid size, 3 mm; scale bars, 100 µm.
A hybrid strategy combining organic and hydrogel clearing
Because rehydration and replacing DBE in FDISCO improved fluorescence but failed to render a whole leg fully transparent, we reasoned that stronger but non-quenching clearing steps are needed for large, heterogeneous, FDISCO-processed tissues to be cleared (Fig. 1g,h). Aqueous methods preserve fluorophores better but have been developed in parallel to DISCO methods and are not thought to be compatible with them (Extended Data Fig. 1a). After realizing that the fluorescence signal was still near 100% after dichloromethane treatment in FDISCO (Fig. 1f), we reasoned that a mild aqueous detergent could be used to further delipidate large samples. However, directly exposing rehydrated FDISCO samples to sodium dodecyl sulfate (SDS) resulted in disintegration of tissues and severe protein loss (Fig. 1i–j), suggesting that additional reinforcement is necessary. Hence, we embedded rehydrated, FDISCO-cleared tissue in CLARITY hydrogel (1% acrylamide, 4% paraformaldehyde) before SDS delipidation (Fig. 1g). Indeed, sequential DISCO delipidation and CLARITY reinforcement improved transparency (Fig. 1k,l) and fluorescence intensity (about tenfold higher than FDISCO, Extended Data Fig. 2a,b) while preserving structural integrity and tissue size and preventing SDS-induced protein loss (Fig. 1i,j and Extended Data Fig. 2c–f), in comparison with the parental methods. By combining the solvent strength of DISCO methods and the reinforcement of polymer-based scaffolding, large heterogeneous tissue samples can be cleared with brief organic treatment followed by detergent clearing, which we have named the HYBRiD protocol.
HYBRiD clearing capability and fluorescence compatibility
In addition to its parental methods, we compared HYBRiD with recently developed methods for large tissue clearing, including uDISCO5, PEGASOS22, and SHANEL24. All tested methods readily cleared the brain (Fig. 2a). Passive clearing alone (without continuous perfusion) was sufficient for SHANEL, uDISCO, and HYBRiD to clear a whole leg (Extended Data Fig. 3a). Next, we evaluated tdTomato fluorescence retention (Fig. 2a,b and Extended Data Fig. 3a,b). In our tests, SHANEL exhibited the highest overall clearing power, however it was designed to be used with immunostaining24; thus, as expected, it severely quenched tdTomato signals. uDISCO, as previously reported6, was incompatible with tdTomato and resulted in weaker fluorescence intensity compared with HYBRiD. Lastly, PEGASOS yielded an intermediate fluorescence intensity but failed to clear a whole leg without perfusion.
a, Representative backlit (top) and confocal (bottom) images of 1-mm-thick PV-Ai9 brain slices processed by the indicated protocols. Shown are tdTomato+ neurons in the neocortex from six samples per condition. Contrast-enhanced (eightfold) images are shown in insets. HYBRiD samples were imaged with one-fourth the laser power used in other methods to avoid oversaturation. DPE, diphenyl ether. b, Quantification of tdTomato fluorescence intensity for HYBRiD (n = 29), uDISCO (n = 22), PEGASOS (n = 28), and SHANEL (n = 24). n indicates the number of measurements from four samples per condition. Statistical significance was determined by one-way ANOVA. c,d, Comparison of HYBRiD and uDISCO with regard to preservation of GFP fluorescence. Representative images (c) and quantification (d) of GFP fluorescence intensity in Thy1-GFP-M brain slices cleared by HYBRiD (n = 40) or uDISCO (n = 24). n indicates the total number of measurements from five samples per condition. Statistical significance was determined by two-tailed t-test. e, Representative images of forelimbs of Thy1-GFP-M mice cleared by HYBRiD or uDISCO, from four samples per condition. f,g, Comparison of HYBRiD and uDISCO with regard to preservation of YFP fluorescence. Representative images (f) and quantification (g) of YFP fluorescence intensity in brain slices cleared by HYBRiD (n = 34) or uDISCO (n = 32). n indicates the number of measurements from five samples per condition. Statistical significance was determined by two-tailed t-test. h, Representative images of forelimb from Thy1-YFP-H mice cleared by HYBRiD or uDISCO from four samples per condition. Grid size, 3 mm; scale bars, 100 µm.
We also tested whether HYBRiD maintained green fluorescent protein (GFP) and yellow fluorescent protein (YFP) signals using brains and limbs from Thy1-GFP (M line) and Thy1-YFP (H line) mice, two models widely used to evaluate clearing compatibility with sparsely expressed fluorophores. Similar to what has been reported for FDISCO6, HYBRiD better preserved both GFP and YFP than did uDISCO under comparable passive clearing conditions (Fig. 2c–h and Extended Data Figs. 2a and 3c–h). We obtained similar results using newborn PV-Ai9 mice (Extended Data Fig. 4a–e).
HYBRiD reveals transgenic fluorophores in whole animals
Transgenic animals are widely used to target and manipulate specific cell types. However, potential 'off-target' leakage of transgene expression (for example, Cre recombinase or fluorophores) has been a long-standing challenge for the development and application of transgenic models. Comprehensive profiling and identification of transgene expression across all organ systems are crucial to interpret phenotypes of genetic mouse models accurately.
For example, the aP2-Cre mouse was initially designed as an adipocyte-specific driver, but it was later revealed that this mouse line also has leaky Cre expression in macrophages and the lymphatic system, confounding a decade of diabetes research using this mouse model25. Traditionally, researchers relied on histology or tissue PCR to detect transgene expression in mouse lines, but this can be tedious and unreliable, especially for cell types that penetrate many different tissues and organs (such as blood cells and nerves). After establishing the HYBRiD protocol, we next tested whether this method could be used to efficiently profile whole-body Cre expression. PV-Cre and somatostatin (SST)-Cre mice have been used to target interneuron populations in the brain, but Cre expression in the rest of the body is less characterized. We applied HYBRiD to whole newborn PV-Ai9 and SST-Ai9 mice and examined the bodies by fluorescent lightsheet microscopy (Fig. 3a). HYBRiD outperformed the parental methods for achieving transparency in a newborn animal (Fig. 3b,c and Extended Data Fig. 4). This allowed 3D profiling of Cre-expressing cells across the body at high resolution (Fig. 3d–g and Supplementary Video 1). In PV-Cre animals, we identified tdTomato+ cells in the cochlea and vestibular system of the inner ear (Fig. 3e), neurons in the dorsal root ganglia (DRG) (Fig. 3f), epithelial cells of the kidneys, wrapping the pulmonary vasculature, and in the colon and bladder (Fig. 3g). Furthermore, we observed distinct patterns of SST-Cre expression in the inner ear, DRGs, kidneys, colon, and bladder (Fig. 3h–k).
a, Workflow of HYBRiD whole-body clearing of newborn mice. b, 3D lightsheet image volume of a newborn mouse after HYBRiD (12 × 12 × 10 mm). In-plane resolution, 1.75 µm; z-step, 4 µm. c, Schematic showing digital slicing planes on 3D volume image to collect images in d and h. d, Digital slice views (500 µm) of a PV-Ai9 cleared newborn mouse in the planes shown in c, taken from 3 samples. Clear structures throughout dorsal to ventral regions are shown. Scale bars: transverse, 1 mm; sagittal, 4 mm; coronal, 2 mm. e, 3D block of inner ear of PV-Ai9 mouse showing detailed structures of the cochlea (vb, vestibular bodies; sg, spiral ganglia). f, Slice view of PV-Ai9 mouse showing expression in DRG neurons and intermediate spinal cord pathways (sc, spinal cord). g, PV expression in multiple abdominal organs and pulmonary vasculature. h–k, The equivalent structures in SST-Ai9 newborn mice as in d–g from three samples. Scale bars, 1 mm unless specified.
We further tested HYBRiD by clearing whole mice at different developmental stages. By tuning the duration of clearing steps, we found that HYBRiD cleared 2-week- and 3-week-old PV-Ai9 juveniles while preserving tdTomato fluorescence (Fig. 4a–f). The cleared bodies can be imaged at single-cell resolution (lateral resolution, 1.75 µm/pixel; z-step, 4 µm) to visualize the whole chest (23 × 20 × 14 mm) using a commercial lightsheet microscope (Fig. 4c,f and Extended Data Fig. 5a–n), or as an entire torso (50 × 30 × 14 mm) with the help of a customized sample chamber (Fig. 4g–q and Extended Data Fig. 5o,p). Fluorescence signals were well preserved throughout the body over a 14-mm imaging depth (>3,500 z-steps), revealing detailed anatomical structures both in coronal views (in-plane) and transverse views (reconstructed x–z planes, Fig. 4h–j and Supplementary Video 2). Notably, we resolved individual DRG neurons (Fig. 4m,n) and renal cortex structures (Fig. 4p,q) in deep tissue >10 mm from the imaging surface (Supplementary Video 3).
a–f, Representative images of 2-week-old (a–c) and 3-week-old (d–f) PV-Ai9 mice before (a and d) and after HYBRiD clearing (b and e) from 4 samples. 3D lightsheet image volumes (as MIP) of the outlined chest areas are shown in c and d. D, dorsal; V, ventral; S, superior; I, inferior; M, medial; L, lateral. g–j, MIP of 3D lightsheet image volume of the 3-week-old PV-Ai9 mouse as indicated in the area enclosed in a red rectangle in e (g). Yellow dashed lines indicate where digital transverse sections (h–j) were taken, showing detailed structures of the thymus (t.y.), aorta (ao), atrium (a.), ventricle (v.), lung (Lu.), diaphragm (d.p.), spinal cord (sc), and liver (lv.). In g, L and R indicate left and right, respectively. k–q, Representative 3D views of 3-week-old PV-Ai9 mice, showing PV-tdTomato expression in the heart (k), DRG (m), intermediate spinal cord spinal cord (o), and kidney (p). l, Zoomed-in view of the yellow square in k, showing PV-tdTomato expression in the atrium. n, Zoomed-in view of the yellow square in m, showing DRG neurons at single-cell resolution. q, Zoomed-in view of the yellow square in p, showing PV-tdTomato expression in renal cortex (r.c.). Scale bars: g, 2,500 µm; c,f, 2,000 µm.
Visualizing SARS-CoV-2 infection in mouse chests
HYBRiD not only maintains the integrity of individual organs, but also keeps the entirety of organ systems en bloc, making it a suitable tool for studying systemic diseases, such as viral infections. HYBRiD clarified the whole chest cavity of SARS-CoV-2-infected K18-hACE2 transgenic mice without perfusion to remove blood or perfusion of organic solvent (Fig. 5a–d). We were able to immunostain and image the entire adult mouse chest (Fig. 5e–j and Supplementary Video 4), even though a large amount of blood accumulated in the diseased tissues before HYBRiD (Fig. 5b,c). We observed multifocal pulmonary infection in K18-hACE2 mice (Fig. 5f–j). Their wild-type counterparts did not show substantial viral deposition as detected by SARS-CoV-2 nucleoprotein immunostaining (Fig. 5e). The SARS-CoV-2 antigen was predominantly seen in alveolar and airway epithelial cells26,27 but not in the pulmonary vasculature28 (Fig. 5g). To examine whether these foci contain multinucleated regions previously reported in vitro29 and in humans30, we subsequently dissected the lungs of the cleared chest for DAPI staining and reimaged these at higher magnifications (Fig. 5k–n). Areas with high levels of SARS-CoV-2 antigen were characterized by frequent syncytia, which are unique in the K18-hACE2 mice and are absent in the wild-type lungs (Fig. 5m,n and Extended Data Fig. 6). The simplicity of the HYBRiD protocol was important here, as we conducted these experiments in a biosafety level 3 facility in which access to equipment and delicate procedures is limited.
a, Workflow of HYBRiD clearing and immunostaining of wild-type (WT) or K18-hACE2 transgenic mice 5 days post infection (DPI) with SARS-CoV-2. b–d, Representative image of the chest at different stages of HYBRiD clearing from 3 samples per group. e,f, MIP of 3D lightsheet image volume of immunostained wild-type (e) or K18-hACE2 (f) chest (19 mm × 19 mm × 13 mm). g, Slice view (100 µm) from f. White arrows indicate absence of nucleoprotein, yellow arrows indicate clusters of viral signal. h–j, Transverse (h), coronal (i), and sagittal (j) view of the chest of a SARS-CoV-2-infected mouse. b, bronchi; cw, chest wall; pc, pleural cavity; pa, pulmonary artery; pv, pulmonary vein; sc, spinal cord; tao, thoracic aorta; ao, aorta; be, bronchiole; es, esophagus; il, inferior lobe; sl, superior lobe; th, thymus; icm, intercostal muscles; ll, left lung; ri, ribs. k, MIP of 3D lightsheet image volume of a half chest sample from another K18-hACE2 mouse. l, Confocal imaging (×10) of the region in a white square in k. m, Confocal imaging (×40) of the region in a white square in l further stained with DAPI. Yellow arrows show multinucleated cells in K18-hACE2 but not wild-type lungs (shown in n). Grid size, 3 mm in all images.
Discussion
By combining the seemingly contrasting principles that underlie the two main brain-clearing methods, we have developed a versatile clearing method for large body parts and whole juvenile rodents while preserving endogenous fluorescence. HYBRiD is uniquely suitable for screening of mouse models and studying multi-organ interactions across broad fields of biomedical research. It also enables researchers who would like to take advantage of the efficiency of organic clearing but hesitate to use organic solvent on their microscopes. Limited by the size of imaging hardware and practical considerations of clearing time, HYBRiD in its current form is applicable for whole juvenile mice or an adult chest (or similarly sized body parts). However, one could integrate shrinkage-based or more aggressive organic clearing methods (such uDISCO or SHANEL) with polymer reinforcements to further balance fluorescence retention and transparency for even larger samples. Nonetheless, HYBRiD overcomes many practical limitations of the parental organic and hydrogel-based methods and offers a simple and scalable option for routine organ and whole-body imaging and screening. These features could accelerate the dissemination of organ- and body-clearing technologies to the broader biomedical research community beyond neuroscience.
Methods
Animals
Mice were group-housed on a 12-hour light/dark cycle and fed a standard rodent chow diet. Room temperature was kept at 22 °C with humidity between 30–80% (not controlled). The following strains were purchased from The Jackson Laboratory for this study: wild-type C57BL/6J (stock no. 000664), B6.129P2-Pvalbtm1(cre)Arbr/J (stock no. 017320, PV-Cre), B6.Cg-Ssttm2.1(cre)Zjh/J (stock no. 013044, SST-Cre), and B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J (stock no. 007909, Ai9). PV-Cre and SST-Cre lines were crossed with Ai9 animals to generate the PV-Ai9 and SST-Ai9 animals used in this study. B6.Cg-Tg(K18-ACE2)2Prlmn/J (stock no. 034860, K18-hACE2) hemizygotes were used for SARS-CoV-2 infection experiments. B6.Cg-Tg(Thy1-EGFP)MJrs/J (stock no. 007788, Thy1-GFP-M) and B6.Cg-Tg(Thy1-YFP)HJrs/J (stock no. 003782, Thy1-YFP-H) animals were a gift from the Deisseroth lab at Stanford University. Both male and female mice were used for anatomical assays. All experimental protocols were approved by The Scripps Research Institute Institutional Animal Care and Use Committee (animal protocol 18-0001) and were in accordance with the guidelines from the US National Institutes of Health.
Sample collection
For brain, leg, and torso samples, adult PV-Ai9, Thy1-GFP, or Thy1-YFP mice (n = 4–6 per genotype) were heavily anesthetized with isoflurane and then transcardially perfused with ice-cold PBS followed by ice-cold 4% PFA in PBS with 4% sucrose (Electron Microscopy Perfusion Fixative, 1224SK). For juvenile whole-body clearing, littermates at 2 weeks (P12, n = 4) and 3 weeks (P21, n = 3) of age were prepared using the above procedures. For newborn whole-body clearing, PV-Ai9 or SST-Ai9 animals age P0–P3 (n = 4–6 per genotype) were anesthetized by hypothermia, and then the front of the rib cage was removed and animals were euthanized by puncture of the right atrium. All samples were washed with PBS and then placed in 4% PFA (in PBS) for post-fixation overnight at 4 °C. For SARS-CoV-2-infected mice, animals were euthanized at 5 days post infection, and their bodies were then submerged in zinc-formalin for fixation without perfusion at 4 °C for 3 days before transfer from the biosafety level 3 (BSL3) facility. After post-fixation, all samples were washed in 1× PBS at room temperature (RT) for 1 hour 3 times to remove residual PFA. All skin and hair was removed. Intact chest samples were prepared by removal of the chest from the body (diaphragm to cervical spine inside rib cage) using scissors, then the scapula and the brown adipose tissue were trimmed off. One sample from each group (wild type and K18-hACE2) was cut in half after clearing (along the central plane of the spine) to facilitate DAPI staining, while the rest were kept intact for fluorescent lightsheet microscopy.
Decalcification and decoloring
Samples (all except 1-mm brain slices, PEGASOS and SHANEL samples) were decalcified in 10% EDTA/15% imidazole solution (in water) with gentle shaking at 4 °C for 4 days10. Subsequently, the samples were washed in PBS with gentle shaking at RT overnight followed by 1-hour washes 3 times at RT. All samples were decolored using 25% N,N,N′,N′-tetrakis(2-hydroxypropyl)ethylenediamine (Quadrol) in 1× PBS with gentle shaking at 37 °C for 2 days (4 days for 3-week-old juveniles); solution was refreshed after 8 hours, then again at 24 hours. Intact SARS-CoV-2-infected chest samples were decolored for 6 days to aid in the removal of substantial amounts of pigment (heme) left in the tissues because they were not perfused. Subsequently, the samples were washed in PBS with gentle shaking at RT overnight followed by 1-hour washes 3 times at RT. Pretreated tissues were stored in 1× PBS with 0.02% sodium azide at 4 °C until clearing was completed.
CLARITY processing
CLARITY samples were first fixed with 4% PFA (in PBS) and then incubated in A1P4 hydrogel (1% acrylamide, 0.125% Bis, 4% PFA, 0.025% VA-044 initiator (w/v), in 1× PBS) at 4 °C for CLARITY embedding and subsequent passive clearing, as previously described15. Samples were transferred to hydrogel for 48–72 hours to allow monomer diffusion. The samples were degassed with nitrogen and polymerized (4 hours at 37 °C) with gentle agitation. Samples were removed from hydrogel and washed with 20 mM LiOH-Boric buffer, pH 8, containing 6% SDS at 37 °C overnight to remove residual PFA and monomers. Buffer was refreshed the next day, and every 3–4 days following during passive clearing. Passive clearing took place at 37 °C with continuous shaking until samples appeared translucent. After clearing, samples were washed in PBST (0.2% Triton X-100) for 2 hours, then the buffer was refreshed and samples were washed overnight at 37 °C to remove residual SDS. Cleared samples were RI matched (EasyIndex, RI = 1.52, LifeCanvas, or PROTOS23) and incubated at 37 °C for 8 hours (up to 3 days) and then 6–8 hours at RT. A vacuum chamber was used to remove any air bubbles in the cleared samples before mounting and imaging.
iDISCO processing
We used a recent iteration of the iDISCO method, Adipoclear3, as previously described. All procedures were carried out at RT with shaking. Fixed samples were washed in 20%, 40%, 60%, and 80% methanol in H2O/0.1% Triton X-100/0.3 M glycine (B1N buffer, pH 7), and 100% methanol for the wash times outlined below. Whole-mount samples were incubated in 100% methanol overnight at 4 °C before delipidation, with omission of the original peroxide bleaching step to preserve maximum fluorescence signal. Samples were then delipidated with 100% dichloromethane (DCM; Sigma-Aldrich), washed in 100% methanol twice, and then washed in 80%, 60%, 40%, and 20% methanol in B1N buffer. Samples were then washed in PBS/0.1% Triton X-100/0.05% Tween 20/2 mg/ml heparin (PTxwH buffer) for 1 hour 3 times, then overnight.
FDISCO processing
All procedures were carried out at 4 °C with shaking as previously described6. Fixed samples were washed in 50%, 70%, and 80% tetrahydrofuran (THF) in 25% quadrol (in 1× PBS to adjust to pH 9), then 95% THF twice. Timing of organic delipidation washes followed the outline given below, with 3-week-old juvenile whole body samples treated with dichloromethane 3 times. Samples were delipidated with 100% dichloromethane (DCM; Sigma-Aldrich) then washed in 95% THF twice, followed by 80%, 70%, and 50% THF. Samples were then washed in 0.2% PBST for 1 hour then 2 hours at RT, followed by 1× PBS for 1 hour 3 times then overnight at RT to thoroughly wash out any remaining organic solvent.
uDISCO, PEGASOS, and SHANEL processing
In comparison experiments, all procedures were completed as previously described, using passive protocols5,22,24. For uDISCO, brain slices were incubated (for reflective index matching) with BABB-D4, and larger samples (legs and bodies) were incubated with BABB-D15, as suggested by the original paper. For PEGASOS, the hard-tissue protocol was used to clear limbs and newborn samples. For SHANEL, the protocol for passive clearing was followed, with modifications to timing due to the differences in sample size from the original publication, which was geared toward larger tissues. Timing of organic washes followed the outline given below.
Organic solvent protocol timing
Each organic clearing method (iDISCO, FDISCO, uDISCO, PEGASOS, SHANEL, HYBRiD) has provided a wide range of options for the length of the organic solvent washing steps according to sample types and fluorescence labels. To enable a direct and efficient comparison among them, we used a fixed wash duration (applied to both organic dehydration and delipidation) across all tested methods when processing the same type of tissues, as summarized here: 1-mm brain sections, 30 minutes; whole mount limbs, 45 minutes; newborn mouse (P0–P3), 90 minutes; adult mouse chest, 90 minutes; juvenile whole body, 180 minutes.
HYBRiD CLARITY embedding and additional clearing
HYBRiD samples were first processed with the rehydrated FDISCO procedure outlined above before gel embedding. See the CLARITY procedure outlined above. The intact chest samples contained high levels of excess blood, which required an increased duration of passive clearing to reduce. Chest samples were passively cleared for at least 28 days (up to 40 days). Active clearing using a SmartClear machine (LifeCanvas Technologies) could accelerate the clearing, but is not required. To remove all SDS from the tissue, samples were washed with 0.2% PBST at 37 °C with shaking twice for 2 hours then overnight, twice.
Immunostaining
Monoclonal rabbit anti-SARS-CoV nucleoprotein (Sinobiological, 40143-R019)28,29 was directly conjugated to AlexaFluor 647 using the APEX Antibody Labeling Kit (Thermo Fisher Scientific, A10475) to allow single-step detection of viral infection in the cleared chest samples. Intact chest samples were incubated with the conjugated antibody at 37 °C for 4 days in 10% DMSO/0.2% PBST/0.02% sodium azide (1:700 dilution from the direct yield of APEX kit), then for an additional 4 days with an increased concentration (1:350 dilution) to maximize penetration of the antibody. Excess antibody was washed off with 0.2% PBST at 37 °C with shaking twice for 2 hours then overnight twice.
Viral infection
B6.Cg-Tg(K18-ACE2)2Prlmn/J (stock no. 034860, K18-hACE2) hemizygotes and wild-type C57BL/6J control mice were purchased from The Jackson Laboratory. Wild-type mice (n = 3) were used as mock-infected controls, and K18-hACE2 transgenic mice (n = 3) (6 mice in total) were infected for tissue-clearing experiments. All mice were maintained in parallel in a BSL3 facility for SARS-CoV-2 infection until euthanization. Mice were infected intranasally with 1 × 105 PFU of SARS-CoV-2 in a final volume of 50 μl following isoflurane sedation. After viral infection, mice were monitored daily and euthanized at 5 days postinfection by CO2. Overall timeline
Sample type | Fixation (d) | Pretreatment (d) | Organic clearing (d) | Embedding (d) | Aqueous clearinga (d) | Immunostaining (d) | RI match (d) |
|---|---|---|---|---|---|---|---|
1-mm brain sections | 1 | - | 1 | 2 | 7 | - | 1 |
Whole mount limbs | 1 | 6 | 1 | 2 | 14 | - | 2 |
Newborn whole body | 2 | 6 | 2 | 3 | 21 | - | 2 |
Adult chest (without perfusion) | 4 | 10 (4 + 6)b | 2 | 3 | 28 | 8 | 3 |
Juvenile whole body (perfused) | 2 | 8 (4 + 4)b | 3 | 3 | 28 | - | 3 |
aOptional use of active clearing will shorten this step by 25–50%. bBuffer was refreshed after 4 days. | |||||||
Imaging
Light-sheet imaging
Whole body and intact chest images were acquired with a lightsheet microscope (SmartSPIM, LifeCanvas). Samples were mounted using 1% agarose/EasyIndex. Whole-body newborns were mounted with the dorsal side (spine) up, while intact chest samples were mounted dorsal side down (spine at the bottom of the Z-range). Samples were securely mounted to the holder after the mounting gel solidified (~20 minutes at 4 °C). Mounted samples were imaged inside the standard SmartSPIM chamber (90 × 65 × 45 mm) filled with 250 ml of EasyIndex after overnight equilibration. All the samples used in this study can be mounted using the standard chamber; however, for >2-week-old mice, only the chests can be visualized owing to the size limit of the standard chamber. To image the whole body of juvenile mice (2 and 3 weeks old), a custom oversized chamber (170 × 90 × 60 mm) was used and filled with 750 ml of EasyIndex. All images were acquired using a ×3.6, 0.2 NA objective (LifeCanvas), with z-step set to 4 µm, exposure time of 2 ms, and sampling at full resolution (2,048 × 2,048, 1.75/1.75/4 µm XYZ voxel size). Image acquisition was completed with bilateral illumination along the central plane of symmetry within the sample.
Confocal microscopy
Cleared samples were incubated in RI matching medium, then mounted to a glass microscope slide using spacers (1 mm, Sunjin Lab). Spacers were coated with silicone (KWIK-SIL, WPI) for use with samples in organic solvent to prevent degradation of the plastic. Tissues were then imaged with the Olympus FV3000 confocal microscope with one of the following objectives: ×4, 0.28 NA, air (XLFluor, Olympus); ×10, 0.6 NA, water immersion (XLUMPlanFI, Olympus); or ×40, 1.25 NA, silicone oil immersion (UPlanSApo, Olympus).
Image postprocessing
Raw data from the SmartSPIM were destriped and stitched using commercial software from LifeCanvas Technologies. Destriped and stitched images were converted to .ims format for 3D visualization in IMARIS (Bitplane, v9.2.1). Maximum intensity projections were captured as snapshots, and digital slice views were generated using the ortho slicer tool. Movies were captured using the recording feature in IMARIS. Confocal images were viewed and adjusted using Fiji (ImageJ v2.1.0).
Quantifications and image analysis
Transmittance
A 6-mm-diameter biopsy punch was used to cut 1-mm-thick brain slices to fit into the wells of a 96-well plate. For leg transparency measurements, 3-mm-thick cross section blocks were cut from the center of the upper hindleg to minimize tissue heterogeneity between samples; then, the punch was used to trim excess tissue. Samples were placed into wells with the minimum volume of RI matching medium needed to submerge them fully. Absorbance was measured by multiarea (5×5 grid across each well) at each wavelength 450 nm–75 nm with Cytation 3 plate reader (BioTek). The central 9 areas (3 × 3) of reading were used for quantification, as the edge of the wells gave varying results owing to the high viscosity of the RI matching medium. Blank was subtracted from raw absorbance values, then converted to transmittance (each condition n = 5).
Signal profile
Signal profile data were generated using Fiji. After opening a Z-projection (maximum intensity projection (MIP)) of an image, a straight line was drawn across an area including signal and background. The plot profile tool was used, and the signal plot data were recorded. Likewise, the same procedure was applied to areas of only background with no signal present. All signal profile plot data were normalized by dividing signal plot by the background plot values.
Tissue size measurement
Size measurement of linear changes were completed using brightfield images of 1-mm brain slices over a grid from each method in RI matching medium, subtracted from PFA-only slices in PBS. Calculations were completed using the square root of the area size change, as previously described6.
Fluorescence intensity and signal-to-background ratio
Fluorescence intensity and signal-to-background ratio were quantified using FIJI based on previous uDISCO and FDISCO publications5,6. For each image, a maximum intensity projection over 50 μm of a region of interest (ROI) of 150–200 μm × 150–200 μm was generated on the surface of the tissue (within 150 μm from the top). The region of the signal was identified by auto-thresholding, and the rest of the image pixels were designated as region of background. For region of signal and region of background, total area, mean intensity, and sum of intensity were measured by FIJI. Fluorescence intensity was then calculated by \({\mathrm{fluorescence}}\;{\mathrm{intensity}} = \frac{{{{{{\mathrm{sum}}}}}\left( {{{{{\mathrm{signal}}}}}} \right) - {{{{\mathrm{mean}}}}}\left( {{{{{\mathrm{background}}}}}} \right) \times {{{{\mathrm{area}}}}}({{{{\mathrm{signal}}}}})}}{{{{{{\mathrm{area}}}}}({{{{\mathrm{signal}}}}})}}\), and signal-to-background ratio was calculated by \({\mathrm{Signal}}\;{\mathrm{to}}\;{\mathrm{background}}\;{\mathrm{ratio}} = \frac{{{{{{\mathrm{mean}}}}}({{{{\mathrm{signal}}}}})}}{{{{{{\mathrm{mean}}}}}({{{{\mathrm{background}}}}})}}\). For each condition, 8–12 random ROIs in the cortex per tissue sample were quantified, and 4–6 tissue samples per condition were used in each analysis.
Protein loss
Samples treated with FDISCO and iDISCO protocols were evaluated for protein loss dependent on sequential hydrogel embedding in comparison to native and CLARITY embedded tissue with no pretreatment. Half-coronal 1 mm PV-Ai9 brain slices were briefly dabbed on a tissue paper to remove excess moisture before mass of tissue in each condition was recorded. All samples placed into LiOH-Boric SDS buffer following their respective treatments and were placed at 37 °C with shaking to begin passively clearing. On the fourth day of clearing, buffer was sampled from each tube, then fresh clearing buffer was added to each tube before samples were placed back on the 37 °C shaker. Samples continued passively clearing until day 7, when clearing buffer was once again collected. A Pierce BCA Protein Assay Kit (cat no. 23227, Thermo Fisher Scientific) was used for colorimetric quantification of protein content. Total protein loss values are the sum of day 4 and day 7 quantifications.
Statistics and reproducibility
Statistical significance was evaluated with Prism 8 (Graphpad) using t-tests and one- or two-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparison post hoc test. All data presented as individual datapoint or mean ± s.e.m., with n values given in the figure legend as biological replicates.
All fluorescence intensity, transparency, size, and protein-loss experiments were repeated 3 times with n = 6. Newborn clearing experiments were repeated 3 times with n = 3 of each method, and each transgenic line. For juvenile clearing experiments, 3–4 biological replicates were compared at ages P12 (2 weeks) and at P21 (3 weeks); for SARS-CoV-2 samples, infection was performed once with 3 biological replicates in each genotype.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All the numeric data in this study are included in the supplemental information. The raw images are too large for public deposit and will be available upon request from the corresponding author. Source data are provided with this paper.
References
Ertürk, A. et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 7, 1983–1995 (2012).
Renier, N. et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014).
Chi, J., Crane, A., Wu, Z. & Cohen, P. Adipo-Clear: a tissue clearing method for three-dimensional imaging of adipose tissue. J. Vis. Exp. 58271 (2018)
Perin, P., Voigt, F. F., Bethge, P., Helmchen, F. & Pizzala, R. iDISCO+ for the study of neuroimmune architecture of the rat auditory brainstem. Front. Neuroanat. 13, 15 (2019).
Pan, C. et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 13, 859–867 (2016).
Qi, Y. et al. FDISCO: advanced solvent-based clearing method for imaging whole organs. Sci. Adv. 5, eaau8355 (2019).
Cai, R. et al. Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull–meninges connections. Nat. Neurosci. 22, 317–327 (2019).
Susaki, E. A. et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014).
Tainaka, K. et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell 159, 911–924 (2014).
Tainaka, K. et al. Chemical landscape for tissue clearing based on hydrophilic reagents. Cell Rep. 24, 2196–2210.e9 (2018).
Susaki, E. A. et al. Versatile whole-organ/body staining and imaging based on electrolyte-gel properties of biological tissues. Nat. Commun. 11, 1982 (2020).
Chung, K. et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013).
Tomer, R., Ye, L., Hsueh, B. & Deisseroth, K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 9, 1682–1697 (2014).
Treweek, J. B. et al. Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat. Protoc. 10, 1860–1896 (2015).
Ye, L. et al. Wiring and molecular features of prefrontal ensembles representing distinct experiences. Cell 165, 1776–1788 (2016).
Neckel, P. H., Mattheus, U., Hirt, B., Just, L. & Mack, A. F. Large-scale tissue clearing (PACT): technical evaluation and new perspectives in immunofluorescence, histology and ultrastructure. Sci. Rep. 6, 34331 (2016).
Yang, B. et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014).
Park, Y.-G. et al. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat. Biotechnol. 37, 73–83 (2019).
Ku, T. et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 34, 973–981 (2016).
Lee, E. et al. ACT-PRESTO: rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Sci. Rep. 6, 18631 (2016).
Ueda, H. R. et al. Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci. 21, 61–79 (2020).
Jing, D. et al. Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Res 28, 803–818 (2018).
Kim, S.-Y. et al. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc. Natl Acad. Sci. USA 112, E6274–E6283 (2015).
Zhao, S. et al. Cellular and molecular probing of intact human organs. Cell 180, 796–812.e19 (2020).
Lee, K. Y. et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes 62, 864–874 (2013).
Winkler, E. S. et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 21, 1327–1335 (2020).
Yinda, C. K. et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog. 17, e1009195 (2021).
Carossino, M. et al. Fatal neuroinvasion and SARS-CoV-2 tropism in K18-hACE2 mice is partially independent on hACE2 expression. Preprint at bioRxiv https://doi.org/10.1101/2021.01.13.425144 (2021).
Buchrieser, J. et al. Syncytia formation by SARS‐CoV‐2‐infected cells. EMBO J. 39, e106267 (2020).
Bussani, R. et al. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine 61, 103104 (2020).
Acknowledgements
This work was supported by the National Institutes of Health Director’s New Innovator Award DP2DK128800 (L. Y.), National Institute of Diabetes and Digestive and Kidney K01DK114165 (L. Y.), Dana Foundation (L. Y.), and Baxter Foundation (L. Y). Y. W. and Z.P. were supported by the Dorris Scholars fellowship. We thank P. Cohen, A. Patapoutian, V. Augustine, and B. Hsueh for discussion. We are grateful to Z. Wu for advice on iDISCO protocols. We also thank D. Huang, F. Zhao, X. Sun, and D. Burton for their input on immunostaining of the SARS-CoV-2 samples. We thank K. Deisseroth and S. Pak of Stanford University for gifting the Thy1-YFP and Thy1-GFP tissue samples. We are grateful to J. Stirman for the imaging support. We thank all members of the Ye lab and the Dorris Neuroscience Center (especially K. Spencer and F. de Souza Polli) for their support and feedback.
Author information
Authors and Affiliations
Contributions
V. N., Y. W., and L. Y. conceived the project and designed the study. V. N., Y. W., Z. P., W. K., and N. L. performed the experiments and analyzed the data. M. H., N. S., J. T., and A. M. provided samples. L. Y. oversaw the study. V. N., Y. W., and L. Y. wrote the manuscript with input from all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Nicolas Renier and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Nina Vogt was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Development of HYBRiD.
A. Schematic of aqueous versus organic clearing protocols. Yellow: organic phase. Blue: aqueous phase. B. Representative images of 1 mm sagittal PV-Ai9 brain slices processed by indicated clearing protocols from 6 samples per group. EI: EasyIndex; DBE: dibenzyl ether. Grid size: 3 mm. C. Quantification of signal-to-background ratio of TdTomato fluorescence signal in PV-Ai9 brain slices processed by indicated clearing protocols. Native (n = 26), CLARITY (n = 31), FDISCO (EI) (n = 30), FDISCO (n = 12), iDISCO (EI) (n = 35), iDISCO (n = 15). (n indicates numbers of measurements from 6 samples per condition). Statistical significance was determined by two-tailed t-tests. D. Representative signal intensity profiles of FDISCO (left) and iDISCO (right) processed PV-Ai9 brain slices in either organic (DBE) or aqueous (EI) RI matching solution. N = 3 for each condition. E. Representative MIP confocal images of 1 mm PV-Ai9 brain slices processed by the indicated clearing methods from 6 samples per condition. Scale bar: 3 mm.
Extended Data Fig. 2 Comparison of HYBRiD and parental methods.
A. Quantification of fluorescence intensity of 1 mm PV-Ai9 brain slices of Native (n = 26), cleared by HYBRiD (n = 30) or FDISCO (n = 12). (n indicates numbers of measurements from 6 samples per condition). Statistical significance of indicated comparison was determined by one-way ANOVA. B. Quantification of transparency of brain slices. All values are mean ± SEM, N = 4 for each group. No significance between HYBRiD and FDISCO, determined by multiple unpaired t-tests. C. Representative images of brain slices processed by the indicated clearing method from 3 samples per condition after 7 days of SDS treatment. D - E. Quantification of protein loss in brain slices (D) and hindlimb (E) during passive SDS clearing. N = 3 per group. Statistical significance was determined by one-way ANOVA. F. Size changes of brain slices after CLARITY, FDISCO and HYBRiD. No significant size changes were detected between CLARITY and HYBRiD. N = 3 per group. Statistical significance was determined by one-way ANOVA.
Extended Data Fig. 3 Comparison of HYBRiD and organic clearing methods.
A. Representative images of hindlimb from PV-Ai9 adult mice processed by the indicated clearing protocols from 6 samples per condition. RI matching media is labeled. B. Quantification of signal-to-background ratio of TdTomato fluorescence signal in PV-Ai9 brain slices processed by HYBRiD (n = 29), uDISCO (n = 22), PEGASOS (n = 28), SHANEL (n = 24). (n indicates numbers of measurements from 4 samples per condition). Statistical significance was determined by one-way ANOVA. C. Quantification of signal-to-background ratio of GFP fluorescence signal in Thy1-GFP-M brain slices processed by HYBRiD (n = 40) or uDISCO (n = 24) (n indicates numbers of measurements from 5 samples per condition). Statistical significance was determined by two-tailed t-test. D. Representative images of hindlimb from Thy1-GFP-M adult mice cleared by HYBRiD or uDISCO (4 samples per group). E. Representative lightsheet images (3D MIP) of forelimb from Thy1-GFP-M adult mice cleared by HYBRiD or uDISCO. F. Quantification of signal to background ratio of YFP fluorescence signal in Thy1-YFP-H brain slices processed by HYBRiD (n = 34) or uDISCO (n = 32) (n indicates numbers of measurements from 5 samples per condition). Statistical significance was determined by two-tailed t-test. G. Representative images of hindlimb from Thy1-YFP-H adult mice cleared by HYBRiD or uDISCO (4 samples per group). H. Representative lightsheet images (3D MIP) of forelimb from Thy1-YFP-H adult mice cleared by HYBRiD or uDISCO from 4 samples per group. Grid size: 3 mm. Scale bar: 1 mm.
Extended Data Fig. 4 Comparison of HYBRiD and other methods on clearing of newborn mice.
A,B. Representative brightfield images of PV-Ai9 newborn mice processed by indicated clearing methods and RI matched in the indicated media (3 samples per condition). Grid size: 3 mm. C - E. Overview of lightsheet imaging of PV-Ai9 newborn mice cleared by the indicated clearing protocols. 3D volume of cleared PV-Ai9 mice from each clearing protocol is shown in coronal view (C), sagittal view (D), and transverse view (E). Note HYBRiD, SHANEL, and uDISCO sample were imaged fully through the D-V axis. D, dorsal; V, ventral; S, superior; I, inferior; L, lateral; M, medial. Scale bar: 2 mm.
Extended Data Fig. 5 HYBRiD clearing of juvenile mice.
A – G. Overview of lightsheet imaging of 2-week-old PV-Ai9 mice cleared by HYBRiD from 4 samples. Transverse (A) and sagittal (B) views showing the chest and heart. Zoom-in views of multiple tissues including heart (C), DRG (D), kidney (E), lung (F) and thymus (G). Labels: atrium(a.), thymus (t.y.), aorta (ao), ventricle (v.), lung (L.), spinal cord (sc), pulmonary vasculature (p.v.), renal cortex (r.c.), and liver (lv.) H – N. Overview of lightsheet imaging of 3-week-old PV-Ai9 mice cleared by HYBRiD from 3 samples. Transverse (H) and sagittal (I) views showing the chest and heart. Zoom-in views of multiple tissues including heart (J), DRG (K), kidney (L), lung (M) and thymus (N). O – P. 3D volume of lightsheet imaging of 3-week-old PV-Ai9 mice cleared by HYBRiD. D, dorsal; V, ventral; S, superior; I, inferior; L, lateral; M, medial. Scale bar: O: 5,000 𝜇m; P: 3,000 𝜇m; A, B, H, I: 1,500 𝜇m; C, J: 1,000 𝜇m; D, F, G, K, M, N: 200 𝜇m; E, L: 150 𝜇m. Inserts in D and K: 50 𝜇m.
Extended Data Fig. 6 SARS-CoV nucleoprotein in WT and K18-hACE2 lung.
MIP of 40x confocal imaging of anti-SARS-CoV nucleoprotein stained lung from WT (A) or K18-hACE2 (B) (3 samples per group). FOV 3 in (B) shows a region with low viral infection as an internal negative control.
Supplementary information
Supplementary Information
Supplementary step-by-step protocol for HYBRiD.
Supplementary Video 1
3D rendering of a HYBRiD-cleared PV-Ai9 newborn mouse (P2) imaged by a lightsheet microscope (SmartSPIM). White signal was from endogenous tdTomato fluorescence. In-plane resolution, 1.75 µm/pixel; z-step size, 4 µm. Image volume: 13 mm × 13 mm × 10 mm.
Supplementary Video 2
Slice view of HYBRiD-cleared 3-week-old PV-Ai9 mouse (P21) imaged as a whole torso by a lightsheet microscope (SmartSPIM). White signal was from endogenous tdTomato fluorescence. The sample was imaged as coronal view (X–Y, 3,540 in-plane frames). Transverse views were digitally resliced from the 3D volumes.
Supplementary Video 3
3D rendering of a HYBRiD cleared 3-week-old PV-Ai9 mouse (P21) imaged as a whole torso by a lightsheet microscope (SmartSPIM). White signal was from endogenous tdTomato fluorescence. In-plane resolution 3.5 µm/pixel (2× downsampled to enable 3D movie rendering), z-step size, 4 µm, 3,540 steps. Image volume: 50 mm × 30mm × 14mm.
Supplementary Video 4
3D rendering of a HYBRiD-cleared chest from a SARS-CoV-2-infected K18-hACE2 transgenic mouse. Blue is the autofluorescence signal used to outline the organs. Red is the anti-SARS-CoV NP AlexaFluor 647 signal. In-plane resolution, 1.75 µm/pixel; z-step size, 4 µm. Image volume: 19 mm × 19 mm × 13 mm.
Source data
Source Data Fig. 1
Statistical Source Data.
Source Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 1
Statistical Source Data.
Source Data Extended Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 3
Statistical Source Data.
Rights and permissions
About this article
Cite this article
Nudell, V., Wang, Y., Pang, Z. et al. HYBRiD: hydrogel-reinforced DISCO for clearing mammalian bodies. Nat Methods 19, 479–485 (2022). https://doi.org/10.1038/s41592-022-01427-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-022-01427-0
This article is cited by
-
TESOS: an integrated approach for uniform mesoscale imaging
Cell Research (2024)
-
Whole-body cellular mapping in mouse using standard IgG antibodies
Nature Biotechnology (2024)
-
Mapping of individual sensory nerve axons from digits to spinal cord with the transparent embedding solvent system
Cell Research (2024)
-
Vagal sensory neurons mediate the Bezold–Jarisch reflex and induce syncope
Nature (2023)
-
Expandable ELAST for super-resolution imaging of thick tissue slices using a hydrogel containing charged monomers
Scientific Reports (2023)