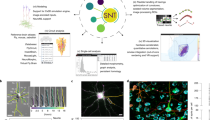
Abstract
Due to advances in automated image acquisition and analysis, whole-brain connectomes with 100,000 or more neurons are on the horizon. Proofreading of whole-brain automated reconstructions will require many person-years of effort, due to the huge volumes of data involved. Here we present FlyWire, an online community for proofreading neural circuits in a Drosophila melanogaster brain and explain how its computational and social structures are organized to scale up to whole-brain connectomics. Browser-based three-dimensional interactive segmentation by collaborative editing of a spatially chunked supervoxel graph makes it possible to distribute proofreading to individuals located virtually anywhere in the world. Information in the edit history is programmatically accessible for a variety of uses such as estimating proofreading accuracy or building incentive systems. An open community accelerates proofreading by recruiting more participants and accelerates scientific discovery by requiring information sharing. We demonstrate how FlyWire enables circuit analysis by reconstructing and analyzing the connectome of mechanosensory neurons.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
FlyWire’s EM data and unproofread segmentation are publicly available. FlyWire’s proofread segmentation is available to the community first as outlined in FlyWire’s principle. Published proofread neurons are publicly available. FlyWire’s website (flywire.ai) describes how to access these different data sources.
All neuron reconstructions used in this manuscript are available and linked in Supplementary Table 2. Additionally, all data necessary to reproduce the analyses in this manuscript are available through the data analysis GitHub repository at https://github.com/seung-lab/FlyWirePaper. This includes the connectivity map between all neurons included in the mechanosensory analyses.
For the comparison with FlyCircuit neurons we used the dotprops of a public dataset60 (https://zenodo.org/record/5205616). Source data are provided with this paper.
Code availability
All repositories presented in this manuscript are open-sourced and available through the Seung laboratory GitHub project. Specifically, our implementation of the ChunkedGraph is available at https://github.com/seung-lab/PyChunkedGraph. Further, the code to reproduce all figures in this manuscript is also available on GitHub at https://github.com/seung-lab/FlyWirePaper.
References
Ahrens, M. B., Orger, M. B., Robson, D. N., Li, J. M. & Keller, P. J. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods 10, 413–420 (2013).
White, J. G., Southgate, E., Thomson, J. N. & Brenner, S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340 (1986).
Cook, S. J. et al. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571, 63–71 (2019).
Scheffer, L. K. et al. A connectome and analysis of the adult Drosophila central brain. eLife https://doi.org/10.7554/eLife.57443 (2020).
Coen, P. et al. Dynamic sensory cues shape song structure in Drosophila. Nature 507, 233–237 (2014).
Duistermars, B. J., Pfeiffer, B. D., Hoopfer, E. D. & Anderson, D. J. A brain module for scalable control of complex, multi-motor threat displays. Neuron 100, 1474–1490 (2018).
Seelig, J. D. & Jayaraman, V. Neural dynamics for landmark orientation and angular path integration. Nature 521, 186–191 (2015).
DasGupta, S., Ferreira, C. H. & Miesenböck, G. FoxP influences the speed and accuracy of a perceptual decision in Drosophila. Science 344, 901–904 (2014).
Owald, D. et al. Activity of defined mushroom body output neurons underlies learned olfactory behavior in Drosophila. Neuron 86, 417–427 (2015).
Zheng, Z. et al. A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell 174, 730–743 (2018).
Kim, J. S. et al. Space-time wiring specificity supports direction selectivity in the retina. Nature 509, 331–336 (2014).
Haehn, D. et al. Design and evaluation of interactive proofreading tools for connectomics. IEEE Trans. Vis. Comput. Graph. 20, 2466–2475 (2014).
Knowles-Barley, S. et al. RhoanaNet pipeline: dense automatic neural annotation. Preprint at arXiv http://arxiv.org/abs/1611.06973 (2016).
Zhao, T., Olbris, D. J., Yu, Y. & Plaza, S. M. NeuTu: software for collaborative, large-scale, segmentation-based connectome reconstruction. Front. Neural Circuits 12, 101 (2018).
Felsenberg, J. et al. Integration of parallel opposing memories underlies memory extinction. Cell 175, 709–722 (2018).
Dolan, M.-J. et al. Communication from learned to innate olfactory processing centers is required for memory retrieval in Drosophila. Neuron 100, 651–668 (2018).
Zheng, Z. et al. Structured sampling of olfactory input by the fly mushroom body. Preprint at bioRxiv https://doi.org/10.1101/2020.04.17.047167 (2020).
Li, P. H. et al. Automated reconstruction of a serial-section EM Drosophila brain with flood-filling networks and local realignment. Preprint at bioRxiv https://doi.org/10.1101/605634 (2019).
Mitchell, E., Keselj, S., Popovych, S., Buniatyan, D. & Sebastian Seung, H. Siamese encoding and alignment by multiscale learning with self-supervision. Preprint at arXiv http://arxiv.org/abs/1904.02643 (2019).
Deutsch, D. et al. The neural basis for a persistent internal state in Drosophila females. eLife https://doi.org/10.7554/eLife.59502 (2020).
Schlegel, P. et al. Information flow, cell types and stereotypy in a full olfactory connectome. eLife https://doi.org/10.7554/eLife.66018 (2021).
Baker, C. A., McKellar, C., Nern, A. & Dorkenwald, S. Neural network organization for courtship song feature detection in Drosophila. Preprint at bioRxiv https://doi.org/10.1101/2020.10.08.332148 (2020).
Pézier, A. P., Jezzini, S. H., Bacon, J. P. & Blagburn, J. M. Shaking B mediates synaptic coupling between auditory sensory neurons and the giant fiber of Drosophila melanogaster. PLoS ONE 11, e0152211 (2016).
Wu, C.-L. et al. Heterotypic gap junctions between two neurons in the Drosophila brain are critical for memory. Curr. Biol. 21, 848–854 (2011).
Lee, K., Zung, J., Li, P., Jain, V. & Sebastian Seung, H. Superhuman accuracy on the SNEMI3D connectomics challenge. Preprint at arXiv https://arxiv.org/abs/1706.00120 (2017).
Dorkenwald, S. et al. Binary and analog variation of synapses between cortical pyramidal neurons. Preprint at bioRxiv https://doi.org/10.1101/2019.12.29.890319 (2019).
Zlateski, A. & Sebastian Seung, H. Image segmentation by size-dependent single linkage clustering of a watershed basin graph. Preprint at arXiv http://arxiv.org/abs/1505.00249 (2015).
Maitin-Shepard, J. et al. google/neuroglancer. Zenodo https://doi.org/10.5281/zenodo.5573294 (2021).
Chang, F. et al. Bigtable: a distributed storage system for structured data. ACM Trans. Comput. Syst. 26, 1–26 (2008).
Priedhorsky, R. et al. Creating, destroying, and restoring value in Wikipedia. in Proc. 2007 International ACM Conference on Supporting Group Work 259–268 https://doi.org/10.1145/1316624.1316663 (2007).
Buhmann, J. et al. Automatic detection of synaptic partners in a whole-brain Drosophila electron microscopy data set. Nat. Methods 18, 771–774 (2021).
Heinrich, L., Funke, J., Pape, C., Nunez-Iglesias, J. & Saalfeld, S. Synaptic cleft segmentation in non-isotropic volume electron microscopy of the complete Drosophila brain. in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018 317–325 (Springer International Publishing, 2018).
Staffler, B. et al. SynEM, automated synapse detection for connectomics. eLife https://doi.org/10.7554/eLife.26414 (2017).
Dorkenwald, S. et al. Automated synaptic connectivity inference for volume electron microscopy. Nat. Methods https://doi.org/10.1038/nmeth.4206 (2017).
Huang, G. B., Scheffer, L. K. & Plaza, S. M. Fully-automatic synapse prediction and validation on a large data set. Front. Neural Circuits 12, 87 (2018).
Turner, N. L. et al. Synaptic partner assignment using attentional voxel association networks. In 2020 IEEE International Symposium on Biomedical Imaging, 1209–1213 (IEEE Computer Society, 2020).
Buhmann, J. et al. in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018 309–316 (Springer International Publishing, 2018).
Kreshuk, A., Funke, J., Cardona, A. & Hamprecht, F. A. in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 661–668 (Springer International Publishing, 2015).
Schneider-Mizell, C. M. et al. Quantitative neuroanatomy for connectomics in Drosophila. eLife https://doi.org/10.7554/eLife.12059 (2016).
Takemura, S.-Y. et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature 500, 175–181 (2013).
Meinertzhagen, I. A. Of what use is connectomics? A personal perspective on the Drosophila connectome. J. Exp. Biol. https://doi.org/10.1242/jeb.164954 (2018).
Chiang, A.-S. et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr. Biol. 21, 1–11 (2011).
Costa, M., Manton, J. D., Ostrovsky, A. D., Prohaska, S. & Jefferis, G. S. X. E. NBLAST: rapid, sensitive comparison of neuronal structure and construction of neuron family databases. Neuron 91, 293–311 (2016).
Tootoonian, S., Coen, P., Kawai, R. & Murthy, M. Neural representations of courtship song in the Drosophila brain. J. Neurosci. 32, 787–798 (2012).
Lai, J. S.-Y., Lo, S.-J., Dickson, B. J. & Chiang, A.-S. Auditory circuit in the Drosophila brain. Proc. Natl Acad. Sci. USA 109, 2607–2612 (2012).
Vaughan, A. G., Zhou, C., Manoli, D. S. & Baker, B. S. Neural pathways for the detection and discrimination of conspecific song in D. melanogaster. Curr. Biol. 24, 1039–1049 (2014).
Yamada, D. et al. GABAergic local interneurons shape female fruit fly response to mating songs. J. Neurosci. 38, 4329–4347 (2018).
Kamikouchi, A. et al. The neural basis of Drosophila gravity-sensing and hearing. Nature 458, 165–171 (2009).
Patella, P. & Wilson, R. I. Functional maps of mechanosensory features in the Drosophila brain. Curr. Biol. 28, 1189–1203 (2018).
Kim, H. et al. Wiring patterns from auditory sensory neurons to the escape and song-relay pathways in fruit flies. J. Comp. Neurol. 528, 2068–2098 (2020).
Clemens, J. et al. Connecting neural codes with behavior in the auditory system of Drosophila. Neuron 87, 1332–1343 (2015).
Azevedo, A. W. & Wilson, R. I. Active mechanisms of vibration encoding and frequency filtering in central mechanosensory neurons. Neuron 96, 446–460 (2017).
von Reyn, C. R. et al. A spike-timing mechanism for action selection. Nat. Neurosci. 17, 962–970 (2014).
Allen, M. J., Godenschwege, T. A., Tanouye, M. A. & Phelan, P. Making an escape: development and function of the Drosophila giant fibre system. Semin. Cell Dev. Biol. 17, 31–41 (2006).
Phelan, P. et al. Molecular mechanism of rectification at identified electrical synapses in the Drosophila giant fiber system. Curr. Biol. 18, 1955–1960 (2008).
Morley, E. L., Steinmann, T., Casas, J. & Robert, D. Directional cues in Drosophila melanogaster audition: structure of acoustic flow and inter-antennal velocity differences. J. Exp. Biol. 215, 2405–2413 (2012).
Giles, J. Internet encyclopaedias go head to head. Nature 438, 900–901 (2005).
Mu, S. et al. 3D reconstruction of cell nuclei in a full Drosophila brain. Preprint at bioRxiv https://doi.org/10.1101/2020.04.17.047167 (2021).
Zung, J. et al. An error detection and correction framework for connectomics. In Proceedings of the 31st International Conference on Neural Information Processing Systems (2017).
Costa, M., Schlegel, P. & Jefferis, G. FlyCircuit Dotprops. Zenodo https://doi.org/10.5281/zenodo.5205616 (2016).
Bates, A. S. et al. The natverse, a versatile toolbox for combining and analysing neuroanatomical data. eLife https://doi.org/10.7554/elife.53350 (2020).
Acknowledgements
We acknowledge support from National Institutes of Health (NIH) BRAIN Initiative RF1 MH117815 to H.S.S. and M.M. M.M. further received funding through an HHMI Faculty Scholar award and an NIH R35 Research Program Award. H.S.S. also received NIH funding through RF1MH123400, U01MH117072, U01MH114824. H.S.S. also acknowledges support from the Mathers Foundation, as well as assistance from Google and Amazon. These companies had no influence on the research. We are grateful for support with FAFB imagery from S. Saalfeld, E. Trautman and D. Bock. We are grateful to D. Bock and Z. Zheng for discussions about FAFB. We thank G. Jefferis, D. Bock, A. Cardona, A. Seeds, S. Hampel and R. Wilson for advice regarding the community. We thank G. Jefferis and P. Schlegel (both with Medical Research Council Laboratory of Molecular Biology and University of Cambridge) for help with the brain renderings, transformations to FlyCircuit and NBLAST comparison with FlyCircuit neurons. We thank G. McGrath for computer system administration and M. Husseini for project administration. We are grateful to J. Maitin-Shepard for Neuroglancer. We are grateful to J. Buhmann and J. Funke for discussions about their synapse resource. We thank N. da Costa, A. Bodor, C. David and the Eyewire team for feedback on the proofreading system. We thank the Allen Institute for Brain Science founder, P. G. Allen, for his vision, encouragement and support. This work was also supported by the Intelligence Advanced Research Projects Activity via Department of Interior/Interior Business Center contract no. D16PC0005 to H.S.S. The US Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright annotation thereon. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of Intelligence Advanced Research Projects Activity, Department of Interior/Interior Business Center or the US Government.
Author information
Authors and Affiliations
Contributions
T.M. and N.K. realigned the dataset with methods developed by E.M., B.N. and T.M. and infrastructure developed by S.P., Z.J. J.A.B., S.M. wrote code for masking defects and misalignments. K.L. trained the convolutional net for boundary detection, using ground-truth data realigned by D.I. J.W. used the convolutional net to generate an affinity map that was segmented by R.L. S.D. and N.K. created the proofreading system with input from J.Z. and Z.A. N.K., M.A.C., O.O., A.H., C.S.J., K.K. and A.R.S. adapted and improved Neuroglancer for proofreading and annotations. S.D., F.C., C.S.M., C.S.J. and D. Brittain built the server infrastructure to host FlyWire and manage users. W.M.S. added the images into cloud storage. C.E.M. managed the community and trained proofreaders. C.E.M., C.J. and A.R.S. designed the training tutorials. C.E.M., C.B., J.G., D.D., L.E.R., S.K., A.B., J.H., M.M., S.M., B.S., K.W., R.W. and D. Bland tested the site and proofread neurons. C.E.M. and J.G. devised neuron annotation procedures. S.C.Y. managed proofreaders and evaluated twigs and synapses. S.D. evaluated the proofreading system. S.D. and C.E.M. analyzed the data. S.D., C.E.M., H.S.S. and M.M. wrote the manuscript. H.S.S. and M.M. led the effort.
Corresponding authors
Ethics declarations
Competing interests
T.M. and H.S.S. are owners of Zetta AI LLC, which provides neural circuit reconstruction services for research laboratories. R.L. and N.K. are employees of Zetta AI LLC.
Additional information
Peer review information Nature Methods thanks Ann-Shyn Chiang, Scott Emmons and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Nina Vogt was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Full brain rendering and comparison with the hemibrain.
(a, b) A neuropil rendering of the fly brain (white) is overlaid with a rendering of the hemibrain and proofread reconstructions of neurons from the antennal mechanosensory and motor center (AMMC). The proofread reconstructions of (a) the AMMC-A2 neuron from the right hemisphere and (b) an WV-WV neuron are added. Scale bar: 50 μm.
Extended Data Fig. 2 Quality of EM image alignment.
(a, b) Chunked pearson correlation (CPC) between two neighboring sections in the original alignment (v14) and our realigned data (v14.1). (a) Relative change of CPC between the original and our realigned data per section. (b) Histogram of the CPC improvements from (a) (dashed red line is at 0). (c, d, e) Example images used for the CPC calculation in (a) where (c) the CPC improved through a better alignment around an artifact, (d) the CPC is almost identical and (e) the CPC overall improved due to a stretch of poorly aligned sections in the original data that were resolved in v14.1.
Extended Data Fig. 3 Chunking the dataset.
(a) Automated segmentation overlayed on the EM data. Each different color represents an individual putative neuron. (b) The underlying supervoxel data is chunked (white dotted lines) such that each supervoxel is fully contained in one chunk. (c) A close up view of the box in (b). (d) Application of the same chunking scheme to the meshes, requiring only minimal mesh recomputations after edits. (e) Diversity of the number of supervoxels in each chunk (median: 25661). (f) The median supervoxel contains 792 voxels. Most very small supervoxels (< 200 voxels) are the result of chunking.
Extended Data Fig. 4 Proofreading with the ChunkedGraph.
(a,)In the ChunkedGraph connected component information is stored in an octree structure where each abstract node (black nodes in levels >1) represents the connected component in the spatially underlying graph (dashed lines represent chunk boundaries). Nodes on the highest layer represent entire neuronal components. (b) Edits in the ChunkedGraph (here, a merge; indicated by the red arrow and added red edge) affect the supervoxel graph to recompute the neuronal connected components. (c) The same neuron shown in Fig. 2 after proofreading with each merged component shown in a different color. Scale bar (c): 10 μm.
Extended Data Fig. 5 The FlyWire proofreading platform.
(a) The most common view in FlyWire displays four panels: a bar with links and a leaderboard of top proofreaders (left), the EM image in grayscale overlaid with segmentation in color (second panel from left), a 3D view of selected cell segments (third panel), and menus with multiple tools (right). (b) Annotation tools include points, which can be used for a variety of purposes such as marking particular cells or synapses.
Extended Data Fig. 6 Fast proofreading in FlyWire.
Analysis of 60 neurons included in the triple proofreading analysis and fast proofreading analysis. (a) Comparison of the F1-Scores (0-1, higher is better; with respect to proofreading results after three rounds) between different proofreading rounds according to volumetric completeness (medians: Auto: 0.777, 1: 0.992, 2: 0.999, Fast: 0.988 means: Auto: 0.729, 1: 0.975, 2: 0.992, Fast: 0.968) and (b) assigned synapses (medians: Auto: 0.799, 1: 0.992, 2: 0.999, Fast: 0.988, means: Auto: 0.746, 1: 0.958, 2: 0.986, Fast: 0.945). ‘Auto’ refers to reconstructions without proofreading. Boxes are interquartile ranges (IQR), whiskers are set at 1.5 x IQR.
Extended Data Fig. 7 NBLAST-based analysis of segmentation accuracy.
Comparison of NBLAST matches and scores of 183 neurons before and after proofreading to assess the quality of the automated segmentation. (a) NBLAST scores of all 183 triple-proofread neurons (Fig. 5) against 16129 neurons in FlyCircuit. For each neuron in FlyWire we found the best hit in FlyCircuit according to the mean of the two NBLAST scores. (b) scores for the best matches labeled by manual labels of match vs. no match (N(match)=174 out of 183). (c) mean scores of the FlyWire neurons with matches before and after proofreading (N = 174 neurons). (d) Histogram of the change in NBLAST score before and after proofreading. (e) Rankings of each FlyCircuit neuron matched to a triple-proofread neuron in FlyWire among the 16129 neurons before proofreading and after one round of proofreading. (f) NBLAST scores of the unproofread segments grouped by whether they matched or did not match the broad cell type after proofreading.
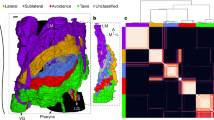
Extended Data Fig. 8 Renderings of AMMC-B1 subtypes.
Neurons grouped by subtype and hemisphere. AMMC, WED brain regions are shown for reference. The neuropil mesh is shown to the same scale. Scale bar: 50 μm.
Extended Data Fig. 9 Connectivity diagrams.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, Supplementary Fig. 1 and Supplementary Note 1
Supplementary Video 1
Introductory Video to FlyWire.
Source data
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
About this article
Cite this article
Dorkenwald, S., McKellar, C.E., Macrina, T. et al. FlyWire: online community for whole-brain connectomics. Nat Methods 19, 119–128 (2022). https://doi.org/10.1038/s41592-021-01330-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-021-01330-0
This article is cited by
-
Petascale pipeline for precise alignment of images from serial section electron microscopy
Nature Communications (2024)
-
Heterogeneity of synaptic connectivity in the fly visual system
Nature Communications (2024)
-
RoboEM: automated 3D flight tracing for synaptic-resolution connectomics
Nature Methods (2024)
-
Local shape descriptors for neuron segmentation
Nature Methods (2023)
-
Flexible circuit mechanisms for context-dependent song sequencing
Nature (2023)