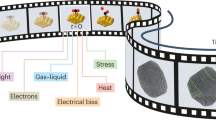
Abstract
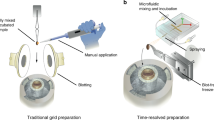
We present an approach for preparing cryo-electron microscopy (cryo-EM) grids to study short-lived molecular states. Using piezoelectric dispensing, two independent streams of ~50-pl droplets of sample are deposited within 10 ms of each other onto the surface of a nanowire EM grid, and the mixing reaction stops when the grid is vitrified in liquid ethane ~100 ms later. We demonstrate this approach for four biological systems where short-lived states are of high interest.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All data and metadata from these studies are available from the corresponding author upon request.
References
Frank, J. Time-resolved cryo-electron microscopy: recent progress. J. Struct. Biol. 200, 303–306 (2017).
Berriman, J. & Unwin, N. Analysis of transient structures by cryo-microscopy combined with rapid mixing of spray droplets. Ultramicroscopy 56, 241–252 (1994).
White, H. D., Walker, M. L. & Trinick, J. A computer-controlled spraying-freezing apparatus for millisecond time-resolution electron cryomicroscopy. J. Struct. Biol. 121, 306–313 (1998).
Unwin, N. & Fujiyoshi, Y. Gating movement of acetylcholine receptor caught by plunge-freezing. J. Mol. Biol. 422, 617–634 (2012).
Subramaniam, S. et al. Protein conformational changes in the bacteriorhodopsin photocycle. J. Mol. Biol. 287, 145–161 (1999).
Lu, Z. et al. Gas-assisted annular microsprayer for sample preparation for time-resolved cryo-electron microscopy. J. Micromech. Microeng. 24, 115001 (2014).
Lu, Z. et al. Passive microfluidic device for sub millisecond mixing. Sens. Actuators B Chem. 144, 301–309 (2010).
Fu, Z. et al. Key intermediates in ribosome recycling visualized by time-resolved cryo-electron microscopy. Structure 24, 2092–2101 (2016).
Jain, T., Sheehan, P., Crum, J., Carragher, B. & Potter, C. S. Spotiton: a prototype for an integrated inkjet dispense and vitrification system for cryo-TEM. J. Struct. Biol. 179, 68–75 (2012).
Dandey, V. P. et al. Spotiton: new features and applications. J. Struct. Biol. 202, 161–169 (2018).
Wei, H. et al. Optimizing ‘self-wicking’ nanowire grids. J. Struct. Biol. 202, 170–174 (2018).
Scapin, G. et al. Structure of the insulin receptor–insulin complex by single-particle cryo-EM analysis. Nature 556, 122–125 (2018).
Zhang, Z. et al. Ensemble cryo-EM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme. eLife 7, e33572 (2018).
Han, H. et al. Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases. eLife 8, e44071 (2019).
Liu, Y. et al. FACT caught in the act of manipulating the nucleosome. Nature 577, 426–431 (2020).
Noble, A. J. et al. Routine single-particle Cryo-EM sample and grid characterization by tomography. eLife 7, e34257 (2018).
Wu, J. L. Y., Tellkamp, F., Khajehpour, M., Robertson, W. D. & Miller, R. J. D. Rapid mixing of colliding picoliter liquid droplets delivered through-space from piezoelectric-actuated pipettes characterized by time-resolved fluorescence monitoring. Rev. Sci. Instrum. 90, 055109 (2019).
Lu, Z. et al. Monolithic microfluidic mixing-spraying devices for time-resolved cryo-electron microscopy. J. Struct. Biol. 168, 388–395 (2009).
Chen, B. et al. Structural dynamics of ribosome subunit association studied by mixing-spraying time-resolved cryogenic electron microscopy. Structure 23, 1097–1105 (2015).
Zadek, B. & Nimigean, C. M. Calcium-dependent gating of MthK, a prokaryotic potassium channel. J. Gen. Physiol. 127, 673–685 (2006).
Posson, D. J., Rusinova, R., Andersen, O. S. & Nimigean, C. M. Calcium ions open a selectivity filter gate during activation of the MthK potassium channel. Nat. Commun. 6, 8342 (2015).
Fan, C. et al. Ball-and-chain inactivation in a calcium-gated potassium channel. Nature 580, 288–293 (2020).
Ruff, E. F., Record, M. T. J. & Artsimovitch, I. Initial events in bacterial transcription initiation. Biomolecules 5, 1035–1062 (2015).
Mazumder, A. & Kapanidis, A. N. Recent advances in understanding σ70-dependent transcription initiation mechanisms. J. Mol. Biol. 431, 3947–3959 (2019).
Saecker, R. M., Record, M. T. J. & Dehaseth, P. L. Mechanism of bacterial transcription initiation: RNA polymerase-promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 412, 754–771 (2011).
Sundborger, A. C. et al. A dynamin mutant defines a superconstricted prefission state. Cell Rep. 8, 734–742 (2014).
Kong, L. et al. Cryo-EM of the dynamin polymer assembled on lipid membrane. Nature 560, 258–262 (2018).
Johansson, M., Bouakaz, E., Lovmar, M. & Ehrenberg, M. The kinetics of ribosomal peptidyl transfer revisited. Mol. Cell 30, 589–598 (2008).
Chen, J. et al. E. coli TraR allosterically regulates transcription initiation by altering RNA polymerase conformation. eLife 8, e49375 (2019).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Sorzano, C. O. S. et al. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J. Struct. Biol. 171, 197–206 (2010).
Lander, G. C. et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009).
Roseman, A. M. FindEM—a fast, efficient program for automatic selection of particles from electron micrographs. J. Struct. Biol. 145, 91–99 (2004).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We are grateful to the staff of the Simons Electron Microscopy Center at the New York Structural Biology Center for help and technical support. We thank I. Fernandez and B. Huang for kindly providing the ribosome subunits. We thank H. He (National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institutes of Health (NIH)) for cryo-EM data collection, NIDDK EM Core Facility and the Hinshaw laboratory for critical comments. The work presented here was conducted at the National Resource for Automated Molecular Microscopy at the New York Structural Biology Center and supported by grants from the NIH (GM103310), the Simons Foundation (SF349247) and the NIDDK NIH Intramural Research Program. Other support included NIH/NIGMS grants R35 GM118130 (S.A.D.) and NIH/RO1GM088352 (C.M.N.).
Author information
Authors and Affiliations
Contributions
V.P.D. and W.C.B. performed all mixing experiments and analyzed data. H.W. assisted with nanowire grid preparation and Spotiton operation. D.B., K.M., M.K. and E.T.E. collected and analyzed cryo-EM data. M.K. assisted with the generation of figures. P.A.K. designed and built the Spotiton system and wrote the operational software. J.E.H. and N.K. contributed sample and biological insights for the dynamin studies. C.M.N., C.F. and N.S. contributed sample and biological insights for the MthK studies. S.A.D., R.M.S., J.C. and B.M. contributed sample and biological insights for the RNAP studies. B.C. and C.S.P. conceived the Spotiton system, designed the experiments and supervised all aspects of the study. V.P.D., W.C.B. and B.C. prepared the manuscript with assistance from all authors.
Corresponding author
Ethics declarations
Competing interests
B.C. and C.S.P. have a commercial relationship with SPT Labtech, a company that produces a commercially available instrument, chameleon, that is based on the Spotiton prototype.
Additional information
Peer review information Arunima Singh was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Specifications of time-resolved Spotiton operation.
a, Diagrammatic overview of the distances (fixed) and elapsed times (variable) relevant to spraying and mixing two samples on a moving grid. Simultaneous dispensing of both samples is triggered after the grid plunge begins. Representative images from the upper and lower cameras are shown directly below the illustrations of each. Sample 1 and sample 2 are indicated in blue and yellow, respectively. b, Magnified view of (green-dashed) boxed area in (a) showing grid and dispensing at specific time-points with corresponding high-speed video captures of the tips and grid below. Elapsed times shown on each image reflect estimates from a video of a grid plunged under Condition 2 (Supplementary Table 1). Objects in (a) and (b) are not drawn to scale. Supplementary Tables 1–4 list values for the following parameters of a grid plunged as depicted in (a) and (b): aaccel, acceleration rate; adecel, deceleration rate; vmax, maximum velocity; t0, plunge start point; tdisp-1, grid leading edge reaches first dispenser; tsamp-1, sample 1 fully applied to grid; tdisp-2, grid leading edge reaches second dispenser; tmix, samples 1 and 2 fully applied to grid; tUC, grid reaches upper camera, tLC, grid reaches lower camera; te, grid plunges into ethane. ‘Spot-to-plunge’ and ‘mix-to-plunge’ in (a) reflect the elapsed times from tdisp-1 or tmix to te, respectively.
Extended Data Fig. 2 Mixing 30 S and 50 S ribosomal subunits to form 70 S complexes.
a, ~20% of particles present were reconstructed to 70 S complex at a resolution of 4.75 Å as indicated by FSC0.5 (b). c, 2D classes of 50 S ribosomal subunit obtained from the control experiment; 2D class of 50S-50S dimer is shown in red. d, 2D classes of the 30 S ribosomal subunit obtained from the control experiment. Both control experiments show no evidence of 70 S ribosomes as observed in the mixed experiment. Scale bars, 20 nm.
Extended Data Fig. 3 Cryo-EM maps of MthK RCK domain with and without Ca2+.
a, The two additional Ca2+ binding sites of MthK either vacant from a control experiment (top row) or occupied after mixing with calcium (bottom row). b, 3D volumes of MthK RCK domains without (top row) and with (bottom row) Ca2+ bound.
Extended Data Fig. 4 Mixing of GTP with dynamin-decorated lipid tubes results in constriction.
Representative cryo-electron micrographs of control dynamin-decorated tubes without GTP (a), with 2 mM GTP (b) and 4 mM GTP (c). Scale bars, 50 nm.
Supplementary information
Supplementary Information
Supplementary Tables 1–4.
Rights and permissions
About this article
Cite this article
Dandey, V.P., Budell, W.C., Wei, H. et al. Time-resolved cryo-EM using Spotiton. Nat Methods 17, 897–900 (2020). https://doi.org/10.1038/s41592-020-0925-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-0925-6
This article is cited by
-
These ‘movies’ of proteins in action are revealing the hidden biology of cells
Nature (2024)
-
Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn2+ uptake into the Golgi apparatus
Nature Communications (2023)
-
A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers
Nature Communications (2023)
-
Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis
Nature Communications (2022)
-
Serial femtosecond crystallography
Nature Reviews Methods Primers (2022)