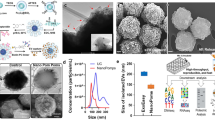
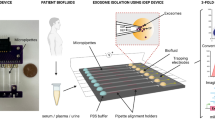
Abstract
Exosomes have shown great potential in disease diagnostics and therapeutics. However, current isolation approaches are burdensome and suffer from low speed, yield and purity, limiting basic research and clinical applications. Here, we describe an efficient exosome detection method via the ultrafast-isolation system (EXODUS) that allows automated label-free purification of exosomes from varied biofluids. We obtained the ultra-efficient purification of exosomes by negative pressure oscillation and double coupled harmonic oscillator–enabled membrane vibration. Our two coupled oscillators generate dual-frequency transverse waves on the membranes, enabling EXODUS to outperform other isolation techniques in speed, purity and yield. We demonstrated EXODUS by purifying exosomes from urine samples of 113 patients and validated the practical relevance in exosomal RNA profiling with the high-resolution capability and high-throughput analysis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request. The raw sequencing data are available at Genome Sequence Archive for Human with Bioproject ID PRJCA003921 and data accession number HRA000457. Source data are provided with this paper.
Code availability
The codes applied in this work are available at Zenodo47 and github website at https://github.com/sterding/EXODUS.
References
Merchant, M. L., Rood, I. M., Deegens, J. K. J. & Klein, J. B. Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nat. Rev. Nephrol. 13, 731–749 (2017).
Wang, W., Luo, J. & Wang, S. Recent progress in isolation and detection of extracellular vesicles for cancer diagnostics. Adv. Healthc. Mater. 7, 1800484 (2018).
Geeurickx, E. & Hendrix, A. Targets, pitfalls and reference materials for liquid biopsy tests in cancer diagnostics. Mol. Asp. Med. 72, 100828 (2020).
Mader, S. & Pantel, K. Liquid biopsy: current status and future perspectives. Oncol. Res. Treat. 40, 404–408 (2017).
Lu, Y. T., Delijani, K., Mecum, A. & Goldkorn, A. Current status of liquid biopsies for the detection and management of prostate cancer. Cancer Manag. Res. 11, 5271–5291 (2019).
Saeedi, S., Israel, S., Nagy, C. & Turecki, G. The emerging role of exosomes in mental disorders. Transl. Psychiat. 9, 122 (2019).
Kanninen, K. M., Bister, N., Koistinaho, J. & Malm, T. Exosomes as new diagnostic tools in CNS diseases. BBA – Mol. Basis Dis. 1862, 403–410 (2016).
Melo, S. A. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523, 177–182 (2015).
Liang, K. et al. Nanoplasmonic quantification of tumor-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat. Biomed. Eng. 1, 0021 (2017).
Li, Y. et al. Circular rna is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25, 981–984 (2015).
Zhu, L. et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer 18, 74 (2019).
Zhang, Z. G., Buller, B. & Chopp, M. Exosomes—beyond stem cells for restorative therapy in stroke and neurological injury. Nat. Rev. Neurol. 15, 193–203 (2019).
Qu, M. et al. Dopamine-loaded blood exosomes targeted to brain for better treatment of Parkinson’s disease. J. Control. Release 287, 156–166 (2018).
Momen-Heravi, F. Isolation of extracellular vesicles by ultracentrifugation. Methods Mol. Biol. 1660, 25–32 (2017).
Li, X. et al. Challenges and opportunities in exosome research-perspectives from biology, engineering, and cancer therapy. APL Bioeng. 3, 011503 (2019).
Guerreiro, E. M. et al. Efficient extracellular vesicle isolation by combining cell media modifications, ultrafiltration, and size-exclusion chromatography. PLoS ONE 13, e0204276 (2018).
Woo, H. K. et al. Exodisc for rapid, size-selective, and efficient isolation and analysis of nanoscale extracellular vesicles from biological samples. ACS Nano 11, 1360–1370 (2017).
Xu, R., Greening, D. W., Zhu, H. J., Takahashi, N. & Simpson, R. J. Extracellular vesicle isolation and characterization: toward clinical application. J. Clin. Invest. 126, 1152–1162 (2016).
Heath, N. et al. Rapid isolation and enrichment of extracellular vesicle preparations using anion exchange chromatography. Sci. Rep. 8, 5730 (2018).
Onódi, Z. et al. Isolation of high-purity extracellular vesicles by the combination of iodixanol density gradient ultracentrifugation and bind-elute chromatography from blood plasma. Front. Physiol. 9, 1479 (2018).
Wu, M. et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl Acad. Sci. USA 114, 10584–10589 (2017).
Li, P., Kaslan, M., Lee, S. H., Yao, J. & Gao, Z. Progress in exosome isolation techniques. Theranostics 7, 789–804 (2017).
Konoshenko, M. Y., Lekchnov, E. A., Vlassov, A. V. & Laktionov, P. P. Isolation of extracellular vesicles: general methodologies and latest trends. Biomed. Res. Int. 2018, 8545347 (2018).
Dalwadi, G., Benson, H. A. E. & Chen, Y. Comparison of diafiltration and tangential flow filtration for purification of nanoparticle suspensions. Pharm. Res. 22, 2152–2162 (2005).
Busatto, S. et al. Tangential flow filtration for highly efficient concentration of extracellular vesicles from large volumes of fluid. Cells 7, https://doi.org/10.3390/cells7120273 (2018).
Colao, I. L., Corteling, R., Bracewell, D. & Wall, I. Manufacturing exosomes: a promising therapeutic platform. Trends Mol. Med. 24, 242–256 (2018).
Andriolo, G. et al. Exosomes from human cardiac progenitor cells for therapeutic applications: development of a GMP-grade manufacturing method. Front Physiol. 9, 1169 (2018).
Fernandez-Cerezo, L. et al. An ultra scale-down method to investigate monoclonal antibody processing during tangential flow filtration using ultrafiltration membranes. Biotechnol. Bioeng. 116, 581–590 (2019).
Subramani, A., DeCarolis, J., Pearce, W. & Jacangelo, J. G. Vibratory shear enhanced process (VSEP) for treating brackish water reverse osmosis concentrate with high silica content. Desalination 291, 15–22 (2012).
Petala, M. D. & Zouboulis, A. I. Vibratory shear enhanced processing membrane filtration applied for the removal of natural organic matter from surface waters. J. Membr. Sci. 269, 1–14 (2006).
Macias, J. D., Ordonez-Miranda, J. & Alvarado-Gil, J. J. Resonance frequencies and Young’s modulus determination of magnetorheological elastomers using the photoacoustic technique. J. Appl. Phys. 112, 124910 (2012).
Soares, R. M. & Gonçalves, P. B. Nonlinear vibrations and instabilities of a stretched hyperelastic annular membrane. Int. J. Solids Struct. 49, 514–526 (2012).
Snowdon, J. C. Forced vibration of damped circular and annular membranes. Tran. Socie. Rheol. 15, 685–707 (1971).
Wolf, M. T. F., Zhang, J. & Nie, M. Uromodulin in mineral metabolism. Curr. Opin. Nephrol. Hypertens. 28, 481–489 (2019).
Lane, R. E., Korbie, D., Hill, M. M. & Trau, M. Extracellular vesicles as circulating cancer biomarkers: opportunities and challenges. Clin. transl. med. 7, 14 (2018).
Boroughs, L. K. & DeBerardinis, R. J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 17, 351–359 (2015).
Sjölund, J. et al. Suppression of renal cell carcinoma growth by inhibition of notch signaling in vitro and in vivo. J. Clin. Invest. 118, 217–228 (2008).
Knowles, M. A. & Hurst, C. D. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat. Rev. Cancer 15, 25–41 (2015).
Marsit, C. J. et al. Epigenetic inactivation of SFRP genes and tp53 alteration act jointly as markers of invasive bladder cancer. Cancer Res. 65, 7081–7085 (2005).
Mateescu, B. et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—an ISEV position paper. J. Extracell. Vesicles 6, 1286095 (2017).
Tracz, A. F., Szczylik, C., Porta, C. & Czarnecka, A. M. Insulin-like growth factor-1 signaling in renal cell carcinoma. BMC Cancer 16, 453 (2016).
Horiguchi, A. et al. Leptin promotes invasiveness of murine renal cancer cells via extracellular signal-regulated kinases and rho dependent pathway. J. Urol. 176, 1636–1641 (2006).
Hanahan, D. & Weinberg, RobertA. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Chen, Y., Zhu, Q., Lee, L. P., & Liu, F. A protocol for exosome detection via the ultrafast-isolation system (EXODUS). Protocol Exchange https://doi.org/10.21203/rs.3.pex-1263/v1 (2020).
Frankish, A. et al. Gencode reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019).
Guo, H. et al. A highly sensitive, self-powered triboelectric auditory sensor for social robotics and hearing aids. Sci. Robot. 3, eaat2516 (2018).
Chen, Y., Zhu, Q., Lee, L. P., & Liu, F. Exosome detection via the ultrafast-isolation system: EXODUS. Zenodo. https://doi.org/10.5281/zenodo.4268892 (2020).
Acknowledgements
We thank Tongji Hospital in the Tongji Medical College at Huazhong University of Science and Technology for providing clinical samples in this study. The work was primarily supported by research fund provided by the Zhenan Technology City Research Fund, the Zhejiang Provincial and Ministry of Health Research Fund for Medical Sciences (grant no. WKJ-ZJ-1910), the Wenzhou Medical University (grant no. 89218012) and the Wenzhou Institute, University of Chinese Academy of Sciences (grant no. WIBEZD2017006-05).
Author information
Authors and Affiliations
Contributions
L.P.L. and F.L. conceived the project and designed the experiments. L.H., L.C. and J.L. organized and collected clinical samples. Y.C. designed the EXODUS device and workstation. Y.C., Q.Z. and Y.W. performed EXODUS system optimization. Y.C., Q.Z., Y.W., M.L. and Q.Y. purified and characterized urinary exosomes. L.H., D.L. and Q.Y. contributed to scanning electron microscopy and TEM analysis of exosomes. Y.W., Q.Y., Q.Z. and M.L. performed isolation method comparison, western blot analysis and NTA for urinary exosomes. Q.Z., Y.W. and Q.Y. performed EV subtype analysis. M.L., Q.Z. and Q.Y. isolated and analyzed saliva samples. M.L., Q.Z. and Q.Y. compared methods for analyzing saliva exosomes. Y.C. and Q.Z. determined the recovery rate. Y.C., Q.Z., M.L., Y.W., L.C., L.H., D.L. and X.D. contributed to data analysis and interpretation. L.C. and J.L. analyzed data related to clinical samples. Y.C. and Q.Z. wrote the manuscript. L.P.L and F.L. edited the manuscript. All experiments were conducted under the supervision of L.P.L. and F.L. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Madhura Mukhopadhyay was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Illustration of EXODUS device.
a, A photo of the EXODUS device, with its cross-sectional illustration. Scale bar, 1 cm. b, Illustration of harmonic oscillations in the EXODUS device for minimizing fouling effect and bio-aggregates. c, The vibration motors generate acoustic streaming at different fluidic levels inside the EXODUS device. Scale bar, 1 cm.
Extended Data Fig. 2 The system design of the EXODUS workstation.
a, An image of the workstation. Scale bar, 10 cm. b, Internal view of the workstation, including (1) autosampler, (2) needle, (3) needle wash site, (4) EXODUS device window, (5) specimen tube, (6) wash buffer A, and (7) wash buffer B. Scale bar, 3 cm. c, The interface to EXODUS device via station: a device slot moves out from inside by pressing the button “Install Device” on the control panel to install EXODUS’s membrane device onto the workstation. d, The architecture of the EXODUS workstation. e, The illustration of the fluidic system in the EXODUS workstation.
Extended Data Fig. 3 Simulations of pressure, flow distribution, and vibration modes of the EXODUS device.
a, COMSOL simulation showing the pressure distribution in a cross-section view of the EXODUS device when applying a −20 kPa negative pressure (NP) from the L-outlet. The sample is injected from the top opening into the sample reservoir. b, Distribution of flow velocity on the membrane from top to bottom. The flow velocity is between 45 and 60 μm/s, with a variation below 25%. c, Simulation of different vibration modes for a clamped circular nanoporous AAO membrane at their representative vibration frequencies: 469.8 Hz (5–700 Hz), 1281.8 Hz (700–1600 Hz), 1861.8 Hz (1600–3000 Hz), 3717.8 Hz (3000–5000 Hz), 5921.8 Hz (5000–8000 Hz), and 10562 Hz (8000–11500 Hz). The integer index m refers to the azimuthal node number. The index n refers to the nth non-trivial zero of the Bessel function. The high-frequency oscillation of the membrane at 6250 Hz by piezoelectric transducer has a (0,4) vibration mode, while the low high-frequency oscillation of the membrane at 200 Hz by vibration motor has a (0,1) vibration mode. The scale ranges from no displacement (dark blue) to maximum displacement (dark red).
Extended Data Fig. 4 Characterization of EXODUS.
a, Comparison of the sample processing times of EXODUS and other methods showing their detailed procedures. For each method, the time cost was basically calculated according to its protocol. b, Size distributions and c amount of the particles obtained by EXODUS and other methods from 10 mL of urine sample aliquots. (n≥5 independent experiments). d, Isolation of exosomes from 10 mL of urine sample aliquots with 20 replications to study the reproducibility of EXODUS. Qubit™ Protein Assay Kits measured the total protein amounts of isolated samples, with an average protein amount of 4.3 μg and a CV of 9.9% over the 20 measurements. e, The concentrations and f size distributions of the particles isolated from different biofluids by EXODUS. (n = 3 independent experiments). The concentration of tear exosomes is presented as particles per centimeter tear collection paper. In c and e, data are presented as mean value ± SD. NTA profiles in b and f are constructed by the average curve (solid line) and error band (shaded area).
Extended Data Fig. 5 NTA and TEM analysis of EV subpopulations.
a, NTA profiles of EV particles with serious cut-off size ranges of 20–450, 20–100, 100–200, and 200–450 nm. b, The typical TEM images of vesicle subpopulations and the corresponding statistics of size distributions are summarized in c. (20–450 nm: n = 565 independent EVs, 20–100 nm: n = 386 independent EVs, 100–200 nm: n = 46 independent EVs, 200–450 nm: n = 31 independent EVs).
Supplementary information
Supplementary Information
Supplementary Figs. 1–17 and Notes 1–3.
Supplementary Video 1
Transverse waves observed at the top surface of liquid in PEI device.
Supplementary Video 2
Transverse waves observed in the deep of PEI device.
Supplementary Video 3
Transverse waves observed in a PMMA device.
Supplementary Table 1
Clinical information of patients
Supplementary Data
Original western blot scans.
Source data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Statistical Source Data
Rights and permissions
About this article
Cite this article
Chen, Y., Zhu, Q., Cheng, L. et al. Exosome detection via the ultrafast-isolation system: EXODUS. Nat Methods 18, 212–218 (2021). https://doi.org/10.1038/s41592-020-01034-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-01034-x
This article is cited by
-
A state-of-the-art review of the recent advances in exosome isolation and detection methods in viral infection
Virology Journal (2024)
-
Efficient preparation of high-purity and intact mesenchymal stem cell–derived extracellular vesicles
Analytical and Bioanalytical Chemistry (2024)
-
Exploiting sound for emerging applications of extracellular vesicles
Nano Research (2024)
-
Nanomaterial Assisted Exosome Analysis Using Mass Spectrometry
Chemical Research in Chinese Universities (2024)
-
Extracellular vesicle-based liquid biopsy biomarkers and their application in precision immuno-oncology
Biomarker Research (2023)