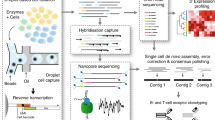
Abstract
We describe droplet-assisted RNA targeting by single-cell sequencing (DART-seq), a versatile technology that enables multiplexed amplicon sequencing and transcriptome profiling in single cells. We applied DART-seq to simultaneously characterize the non-A-tailed transcripts of a segmented dsRNA virus and the transcriptome of the infected cell. In addition, we used DART-seq to simultaneously determine the natively paired, variable region heavy and light chain amplicons and the transcriptome of B lymphocytes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Macosko, E. Z. et al. Cell 161, 1202–1214 (2015).
Klein, A. M. et al. Cell 161, 1187–1201 (2015).
Zheng, G. X. Y. et al. Nat. Commun. 8, 14049 (2017).
Zanini, F., Pu, S.-Y., Bekerman, E., Einav, S. & Quake, S. R. eLife 7, e32942 (2018).
Russell, A. B., Trapnell, C. & Bloom, J. D. eLife 7, e32303 (2018).
Steuerman, Y. et al. Cell Syst. 6, 679–691 (2018).
Parker, J. S., Broering, T. J., Kim, J., Higgins, D. E. & Nibert, M. L. J. Virol. 76, 4483–4496 (2002).
Ooms, L. S., Jerome, W. G., Dermody, T. S. & Chappell, J. D. J. Virol. 86, 10979–10987 (2012).
Georgiou, G. et al. Nat. Biotechnol. 32, 158–168 (2014).
DeKosky, B. J. et al. Nat. Med. 21, 86–91 (2015).
Vollmers, C., Sit, R. V., Weinstein, J. A., Dekker, C. L. & Quake, S. R. Proc. Natl Acad. Sci. USA 110, 13463–13468 (2013).
Kaminski, D. A., Wei, C., Qian, Y., Rosenberg, A. F. & Sanz, I. Front. Immunol. 3, 302 (2012).
Smith, K. et al. Vaccine 34, 2813–2820 (2016).
Abe, M. et al. Clin. Exp. Immunol. 111, 457–462 (1998).
Barandun, S. in Verhandlungen der Deutschen Gesellschaft für innere Medizin. 84. Kongreß (ed. Schlegel, B.) 481–490 (J.F. Bergmann-Verlag, 1978).
Kugelberg, E. Nat. Rev. Immunol. 15, 133 (2015).
Mroczek, E. S. et al. Front. Immunol. 5, 96 (2014).
DeKosky, B. J. et al. Proc. Natl Acad. Sci. USA 113, E2636–E2645 (2016).
DeKosky, B. J. et al. Nat. Biotechnol. 31, 166–169 (2013).
Weinstein, J. A., Zeng, X., Chien, Y.-H. & Quake, S. R. PLoS ONE 8, e67624 (2013).
Saikia, M. et al. Protocol Exchange https://doi.org/10.1038/protex.2018.118 (2018).
Gansauge, M.-T. et al. Nucleic Acids Res. 45, e79 (2017).
Patton, J. T. & Spencer, E. Virology 277, 217–225 (2000).
Joklik, W. K. Microbiol. Rev. 45, 483–501 (1981).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Nat. Biotechnol. 33, 495–502 (2015).
van der Maaten, L. & Hinton, G. E. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Kobayashi, T. et al. Cell Host Microbe 1, 147–157 (2007).
Li, H. et al. Bioinformatics 25, 2078–2079 (2009).
Bolotin, D. A. et al. Nat. Methods 12, 380–381 (2015).
Acknowledgements
We thank P. Schweitzer and colleagues at the Cornell Biotechnology Resource Center (BRC) for help with sequencing assays. This work was supported by US National Institutes of Health (NIH; grant 1DP2AI138242 to I.D.V. and NIAID R01AI121216 to J.S.L.P.) and the National Science Foundation Graduate Research Fellowship Program (NSF-GRFP; grant DGE-1144153 to P.B.).
Author information
Authors and Affiliations
Contributions
P.B., M.S., C.G.D., J.S.L.P. and I.D.V. designed the study. P.B., M.S., S.H.K., M.H., P.M.-L. and M.M.H. carried out the experiments. P.B., M.S., M.F.Z.W. and I.D.V. analyzed the data. P.B., M.S. and I.D.V. wrote the manuscript. All authors provided comments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Protocol for converting Drop-seq primer beads to DART-seq primer beads.
Double-stranded toehold probes with a poly-(dA) ssDNA overhang are annealed to a subset of oligos on the surface of Drop-seq primer beads (left). The toehold is ligated to the bead with T4 DNA ligase. The complementary toehold strand is removed after ligation via heating. DART-seq beads (right) then contain custom primers as well as oligo(dT) tails from the original Drop-seq bead.
Supplementary Figure 2 A fluorescence hybridization assay characterizes the efficiency and tunability of the DART-seq ligation reaction.
a) Assay design to test the efficiency of custom primer ligation to Drop-seq primer beads: (1) DART-seq beads are created withthe addition of custom primers at various concentrations, (2) oligos complementary to the custom primer, and labeled with Cy5, are hybridized to the DART-seq beads, and (3) the fluorescence signal of 3,000 beads is measured with a Qubit 3.0 fluorometer. b) Fluorescence signal as a function of the quantity of Cy5 oligos.
Supplementary Figure 3 DART-seq is reproducible between biological replicates.
Per-base sequence coverage of viral genome segments relative to the number of host transcripts detected, measured by DART-seq and Drop-seq, is shown. Dotted lines indicate the position of the custom primer target sites for DART-seq design 1, and bolded boxes represent the addition of custom primers from DART-seq design 2 (see main text). No viral sequences were detected in a noninfected cell line.
Supplementary Figure 4 DART-seq does not significantly alter the detection of poly(A)-tailed mRNA.
(a-b) Number of UMIs (a) and unique genes (b) detected as a function of cell rank for DART-seq and Drop-seq for the same sample (CD19+ B cells). (c-d) Violin plots of the number of UMIs (c) and genes detected (d) in DART-seq and Drop-seq assays for the same sample (CD19+ B cells). To make these comparisons, we sampled both datasets to the same number of raw sequences (167 × 106). The violin plots show the probability density distributions of the respective variables.
Supplementary Figure 5 DART-seq outperforms Drop-seq in the detection of heavy- and light chain sequences for B cells within human PBMCs.
Sigmoidal plots indicate the percentage of B cells for which heavy- and/or light chain transcripts were detected as a function of the UMI count per cell. Cells were binned by the number of UMIs detected (bin width 200 UMIs, 0–2,400 UMIs per cell, bins with fewer than 20 cells omitted, 26–2,396 cells per bin). Distributions were fit with a sigmoid curve (described in Methods).
Supplementary Figure 6 DART-seq captures a diverse variable region in immune repertoires of B cells.
Comparison of variable isoforms detected with DART-seq (164 cells, PBMC dataset) within Ig heavy and Ig light variable regions. Each column represents a separate, detected variable subtype, normalized by the total number of variable regions detected with respect to light or heavy chains.
Supplementary Figure 7 VH and VL expression in single B cells is correlated.
The fraction of heavy chain (VH) transcripts versus the fraction of variable light chain (VL) transcripts detected in single cells is depicted (PBMC dataset, B cells for which the complete CDR3L and CDR3H region was detected, n = 120). The blue line represents the best fit from linear regression (shaded area represent 95% confidence interval). Pearson correlation 0.683 (P << 10–10).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7
Supplementary Protocol
DART-seq
Supplementary Table 1
Oligonucleotide table
Supplementary Software
Files related to DART-seq including image analysis, viral infection and B-cell-repertoire analysis. These files, as well as images from the image analysis module, are at https://github.com/pburnham50/DART-seq.
Source data
Rights and permissions
About this article
Cite this article
Saikia, M., Burnham, P., Keshavjee, S.H. et al. Simultaneous multiplexed amplicon sequencing and transcriptome profiling in single cells. Nat Methods 16, 59–62 (2019). https://doi.org/10.1038/s41592-018-0259-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0259-9
This article is cited by
-
Single cell RNA-seq: a novel tool to unravel virus-host interplay
VirusDisease (2024)
-
CRISPR screening in hematology research: from bulk to single-cell level
Journal of Hematology & Oncology (2023)
-
Spatial mapping of the total transcriptome by in situ polyadenylation
Nature Biotechnology (2023)
-
Spatial total RNA-sequencing maps coding, noncoding and viral RNAs in tissues
Nature Biotechnology (2023)
-
The Next Generation of Microbial Ecology and Its Importance in Environmental Sustainability
Microbial Ecology (2023)