Abstract
CRISPR/Cas9-based therapies hold considerable promise for the treatment of genetic diseases. Among these, Hutchinson–Gilford progeria syndrome, caused by a point mutation in the LMNA gene, stands out as a potential candidate. Here, we explore the efficacy of a CRISPR/Cas9-based approach that reverts several alterations in Hutchinson–Gilford progeria syndrome cells and mice by introducing frameshift mutations in the LMNA gene.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Code availability
The in-house Perl scripts used for gene edition analysis can be freely downloaded at https://github.com/vqf/genotypes.
Data availability
The MiSeq data were deposited in the NCBI SRA (no. PRJNA505974). Other data from this study are available from the corresponding authors. Any materials that can be shared will be released via a Material Transfer Agreement.
References
Hennekam, R. C. Am. J. Med. Genet. A. 140, 2603–2624 (2006).
De Sandre-Giovannoli, A. et al. Science 300, 2055 (2003).
Eriksson, M. et al. Nature 423, 293–298 (2003).
Goldman, R.D. et al. Proc. Natl Acad. Sci. USA 101, 8963–8968 (2004).
Osorio, F. G. et al. Sci. Transl. Med. 3, 106ra107 (2011).
Fong, L. G. et al. J. Clin. Invest. 116, 743–752 (2006).
Gordon, L. B., Rothman, F. G., Lopez-Otin, C. & Misteli, T. Cell 156, 400–407 (2014).
Gordon, L. B. et al. Circulation 130, 27–34 (2014).
Gordon, L. B. et al. JAMA 319, 1687–1695 (2018).
Amoasii, L. et al. Sci. Transl. Med. 9, eaan8081 (2017).
Yang, Y. et al. Nat. Biotechnol. 34, 334–338 (2016).
Gao, X. et al. Nature 553, 217–221 (2018).
Doudna, J. A. & Charpentier, E. Science 346, 1258096 (2014).
de la Rosa, J. et al. Nat. Commun. 4, 2268 (2013).
Ran, F. A. et al. Nature 520, 186–191 (2015).
Scaffidi, P. & Misteli, T. Nat. Med. 11, 440–445 (2005).
Bar, D. Z. et al. J. Med. Genet. 54, 212–216 (2017).
Gaudelli, N. M. et al. Nature 551, 464–471 (2017).
Beyret, E. et al. Nat. Med. https://doi.org/10.1038/s41591-019-0343-4 (2019).
Li, H. & Durbin, R. Bioinformatics 26, 589–595 (2010).
Li, H. et al. Bioinformatics 25, 2078–2079 (2009).
Acknowledgements
We thank G. Velasco, R. Villa-Bellosta, C. Bárcena, A.P. Ugalde and X.M. Caravia for helpful comments and advice, and R. Feijoo, A. Moyano, D.A. Puente and S.A. Miranda for excellent technical assistance. We also acknowledge the generous support by J.I. Cabrera and Associazione Italiana Progeria Sammy Basso and the contribution of Dr. Matthew Golding to the generation of the anti-murine progerin antibody. The Instituto Universitario de Oncología del Principado de Asturias is supported by Fundación Bancaria Caja de Ahorros de Asturias. J.M.P.F. is supported by Ministerio de Economía y Competitividad (MINECO/FEDER: No. SAF2015-64157-R) and Gobierno del Principado de Asturias. C.L.-O. is supported by grants from the European Research Council (ERC-2016-ADG, DeAge), Ministerio de Economía y Competitividad (MINECO/FEDER: Nos. SAF2014-52413-R and SAF2017-87655-R), Instituto de Salud Carlos III (RTICC) and Progeria Research Foundation (No. PRF2016-66). O.S.-F. is recipient of an FPU fellowship. A.R.F. is recipient of a Ramón y Cajal fellowship. The generation of progerin antibody was funded by the Wellcome Trust (No. 098291/Z/12/Z to S.N.).
Author information
Authors and Affiliations
Contributions
F.G.O., J.M.P.F. and C.L.-O. conceived and designed experiments. O.S.-F., F.G.O., V.Q., F.R., S.B., D.M. and A.R.F. performed experiments and analyzed data. L.R., A.B. and S.N. provided reagents. O.S.-F., F.G.O., A.R.F., J.M.P.F., and C.L.-O. wrote the manuscript. All authors revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
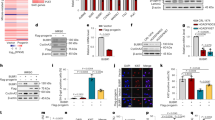
Extended Data Fig. 1 Representative capillary electrophoresis-based fragment analysis of sgRNA-control- and sgRNA-LCS1-transduced LmnaG609G/G609G mouse embryonic fibroblasts (n = 3 independent infections and MEF lines).
Red line and orange peaks correspond to size standards.
Extended Data Fig. 2 Representative capillary electrophoresis-based fragment analysis of sgRNA-control- and sgRNA-LCS1-transduced LMNAG608G/+ human fibroblasts (n = 3 independent infections).
Red line and orange peaks correspond to size standards.
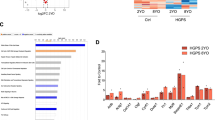
Extended Data Fig. 3 Percentage of indels in LmnaG609G/G609G sgRNA-LC2-transduced male and female mouse tissues.
Data are mean ± s.e.m. (n = 5 tissues per group, except in sgRNA-LCS2-transduced female liver where n = 4; two-tailed Student’s t-test).
Extended Data Fig. 4 RT–qPCR analysis of progerin and lamin C in tissues from LmnaG609G/G609G sgRNA-control-transduced and LmnaG609G/G609G sgRNA-LCS2-transduced mice.
Data are mean ± s.e.m. (n = 4 tissues per group, except sgRNA-control-transduced liver and heart where n = 5; two-tailed Student’s t-test).
Extended Data Fig. 5
Progerin immunohistochemistry in lung, kidney and aorta from WT, LmnaG609G/G609G sgRNA-control-transduced and LmnaG609G/G609G sgRNA-LCS2-transduced mice (lung and kidney, n = 5 for WT and sgRNA-control-transduced mice and n = 4 for sgRNA-LCS2-transduced mice; aorta, n = 2 for WT and n = 3 for sgRNA-control- and sgRNA-LCS2-transduced LmnaG609G/G609G mice). Scale bar, 100 μm.
Extended Data Fig. 6
Kaplan–Meier survival plot of LmnaG609G/G609G male and female mice transduced with sgRNA-control (n = 6 males; n = 4 females) or sgRNA-LCS2 (n = 4 males; n = 6 females).
Extended Data Fig. 7 Progression of body weight of male and female mice transduced with sgRNA-control or sgRNA-LCS2, expressed as percentage of weight at 9 weeks.
Mean values ± s.e.m. are shown (for males, initial n = 9 sgRNA-control-transduced mice and n = 8 sgRNA-LCS2-transduced mice; for females, initial n = 6 mice per group; two-tailed Student’s t-test). Vertical arrow indicates the time point (3.5 months) at which the cohort destined for histological studies was sacrificed.
Extended Data Fig. 8
Images of three sex- and age-matched mice transduced with the sgRNA-LCS2 compared to sgRNA-control-transduced animals.
Extended Data Fig. 9 H&E staining of gastric mucosa from WT, LmnaG609G/G609G sgRNA-control-transduced and LmnaG609G/G609G sgRNA-LCS2-transduced mice.
The graph shows atrophy quantification according to a pathological score as described in Methods. Data are mean ± s.e.m. (n = 5 for WT and sgRNA-control-transduced mice; n = 3 for sgRNA-LCS2-transduced mice).
Supplementary information
Supplementary Video 1
LmnaG609G/G609G sgRNA-control-transduced (top) versus sgRNA-LCS2-transduced (bottom) mice. Both are 4-month-old littermates of the same gender.
Supplementary Tables
Supplementary Tables 1–4
Source data
Source data Fig. 1
Unprocessed Western Blots
Source data Fig. 1
Statistical Source Data
Source Data Fig. 2
Statistical Source Data
Extended Data Source Data Fig. 3
Statistical Source Data
Extended Data Source Data Fig. 4
Statistical Source Data
Extended Data Source Data Fig. 6
Statistical Source Data
Extended Data Source Data Fig. 7
Statistical Source Data
Rights and permissions
About this article
Cite this article
Santiago-Fernández, O., Osorio, F.G., Quesada, V. et al. Development of a CRISPR/Cas9-based therapy for Hutchinson–Gilford progeria syndrome. Nat Med 25, 423–426 (2019). https://doi.org/10.1038/s41591-018-0338-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0338-6
This article is cited by
-
Recent advances in therapeutic CRISPR-Cas9 genome editing: mechanisms and applications
Molecular Biomedicine (2023)
-
Unique progerin C-terminal peptide ameliorates Hutchinson–Gilford progeria syndrome phenotype by rescuing BUBR1
Nature Aging (2023)
-
Progeria—a Rare Genetic Condition with Accelerated Ageing Process
Applied Biochemistry and Biotechnology (2023)
-
Safeguarding genome integrity during gene-editing therapy in a mouse model of age-related macular degeneration
Nature Communications (2022)
-
Transient expression of an adenine base editor corrects the Hutchinson-Gilford progeria syndrome mutation and improves the skin phenotype in mice
Nature Communications (2022)