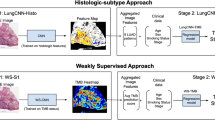
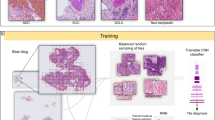
Abstract
Visual inspection of histopathology slides is one of the main methods used by pathologists to assess the stage, type and subtype of lung tumors. Adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC) are the most prevalent subtypes of lung cancer, and their distinction requires visual inspection by an experienced pathologist. In this study, we trained a deep convolutional neural network (inception v3) on whole-slide images obtained from The Cancer Genome Atlas to accurately and automatically classify them into LUAD, LUSC or normal lung tissue. The performance of our method is comparable to that of pathologists, with an average area under the curve (AUC) of 0.97. Our model was validated on independent datasets of frozen tissues, formalin-fixed paraffin-embedded tissues and biopsies. Furthermore, we trained the network to predict the ten most commonly mutated genes in LUAD. We found that six of them—STK11, EGFR, FAT1, SETBP1, KRAS and TP53—can be predicted from pathology images, with AUCs from 0.733 to 0.856 as measured on a held-out population. These findings suggest that deep-learning models can assist pathologists in the detection of cancer subtype or gene mutations. Our approach can be applied to any cancer type, and the code is available at https://github.com/ncoudray/DeepPATH.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All relevant data used for training during the current study are available through the Genomic Data Commons portal (https://gdc-portal.nci.nih.gov). These datasets were generated by TCGA Research Network (http://cancergenome.nih.gov/), and they have made them publicly available. Other datasets analyzed during the current study are available from the corresponding author on reasonable request.
References
Travis, W. D. et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 6, 244–285 (2011).
Hanna, N. et al. Systemic therapy for stage IV non–small-cell lung cancer: American Society of Clinical Oncology clinical practice guideline update. J. Clin. Oncol. 35, 3484–3515 (2017).
Chan, B. A. & Hughes, B. G. Targeted therapy for non–small cell lung cancer: current standards and the promise of the future. Transl. Lung Cancer Res. 4, 36–54 (2015).
Parums, D. V. Current status of targeted therapy in non–small cell lung cancer. Drugs Today (Barc) 50, 503–525 (2014).
Terra, S. B. et al. Molecular characterization of pulmonary sarcomatoid carcinoma: analysis of 33 cases. Mod. Pathol. 29, 824–831 (2016).
Blumenthal, G. M. et al. Oncology drug approvals: evaluating endpoints and evidence in an era of breakthrough therapies. Oncologist 22, 762–767 (2017).
Pérez-Soler, R. et al. Determinants of tumor response and survival with erlotinib in patients with non–small-cell lung cancer. J. Clin. Oncol. 22, 3238–3247 (2004).
Jänne, P. A. et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 14, 38–47 (2013).
Thunnissen, E., van der Oord, K. & den Bakker, M. Prognostic and predictive biomarkers in lung cancer. A review. Virchows Arch. 464, 347–358 (2014).
Zachara-Szczakowski, S., Verdun, T. & Churg, A. Accuracy of classifying poorly differentiated non–small cell lung carcinoma biopsies with commonly used lung carcinoma markers. Hum. Pathol. 46, 776–782 (2015).
Luo, X. et al. Comprehensive computational pathological image analysis predicts lung cancer prognosis. J. Thorac. Oncol. 12, 501–509 (2017).
Yu, K.-H. et al. Predicting non–small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 7, 12474 (2016).
Khosravi, P., Kazemi, E., Imielinski, M., Elemento, O. & Hajirasouliha, I. Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. EBioMedicine 27, 317–328 (2018).
Sozzi, G. et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J. Clin. Oncol. 21, 3902–3908 (2003).
Terry, J. et al. Optimal immunohistochemical markers for distinguishing lung adenocarcinomas from squamous cell carcinomas in small tumor samples. Am. J. Surg. Pathol. 34, 1805–1811 (2010).
Schmidhuber, J. Deep learning in neural networks: an overview. Neural Netw. 61, 85–117 (2015).
Greenspan, H., Ginneken, Bv & Summers, R. M. Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans. Med. Imaging 35, 1153–1159 (2016).
Qaiser, T., Tsang, Y.-W., Epstein, D. & RajpootEma, N. Tumor segmentation in whole slide images using persistent homology and deep convolutional features. In Medical Image Understanding and Analysis: 21st Annual Conference on Medical Image Understanding and Analysis. (Eds. Valdes Hernandez, M. & González-Castro, V.) 320–329 (Springer International Publishing, New York, 2018).
Shen, D., Wu, G. & Suk, H.-I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 19, 221–248 (2017).
Xing, F., Xie, Y. & Yang, L. An automatic learning-based framework for robust nucleus segmentation. IEEE Trans. Med. Imaging 35, 550–566 (2016).
de Bel, T. et al. Automatic segmentation of histopathological slides of renal tissue using deep learning. In Medical Imaging 2018: Digital Pathology Vol. 10581 (Eds. Tomaszewski, J. E. & Gurcan, M. N.) 1058112 (International Society for Optics and Photonics, Bellingham, WA, USA, 2018).
Simon, O., Yacoub, R., Jain, S., Tomaszewski, J. E. & Sarder, P. Multi-radial LBP features as a tool for rapid glomerular detection and assessment in whole slide histopathology images. Sci. Rep. 8, 2032 (2018).
Cheng, J.-Z. et al. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci. Rep. 6, 24454 (2016).
Cruz-Roa, A. et al. Accurate and reproducible invasive breast cancer detection in whole-slide images: a deep learning approach for quantifying tumor extent. Sci. Rep. 7, 46450 (2017).
Sirinukunwattana, K. et al. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans. Med. Imaging 35, 1196–1206 (2016).
Ertosun, M. G. & Rubin., D. L. Automated grading of gliomas using deep learning in digital pathology images: a modular approach with ensemble of convolutional neural networks. In AMIA Annual Symposium Proceedings. 1899–1908 (American Medical Informatics Association, Bethesda, MD, USA).
Bulten, W., Kaa, C.A.H.-d., Laak, J.d. & Litjens, G.J. Automated segmentation of epithelial tissue in prostatectomy slides using deep learning. In Medical Imaging 2018: Digital Pathology. Vol. 10581 (Eds. Tomaszewski, J. E. & Gurcan, M. N.) 105810S (International Society for Optics and Photonics, Bellingham, WA, USA, 2018).
Mishra, R., Daescu, O., Leavey, P., Rakheja, D. & Sengupta, A. Histopathological Diagnosis for Viable and Non-viable Tumor Prediction for Osteosarcoma Using Convolutional Neural Network. In International Symposium on Bioinformatics Research and Applications Vol. 10330 (Eds. Cai, Z., D. Ovidiu, & Li, M.) 12–23 (Springer International Publishing, New York, 2018).
Anthimopoulos, M., Christodoulidis, S., Ebner, L., Christe, A. & Mougiakakou, S. Lung Pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans. Med. Imaging 35, 1207–1216 (2016).
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2818–2826 (Boston, MA, USA, 2015).
Szegedy, C. et al. Going Deeper With Convolutions. In The IEEE Conference on Computer Vision and Pattern Recognition. 1–9 (Boston, 2015).
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017).
Gulshan, V. et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. J. Am. Med. Assoc. 316, 2402–2410 (2016).
Grossman, R. L. et al. Toward a shared vision for cancer genomic data. N. Engl. J. Med. 375, 1109–1112 (2016).
Abels, E. & Pantanowitz, L. Current state of the regulatory trajectory for whole slide imaging devices in the USA. J. Pathol. Inform. 8, 23 (2017).
Sanchez-Cespedes, M. et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 62, 3659–3662 (2002).
Shackelford, D. B. et al. LKB1 inactivation dictates therapeutic response of non–small cell lung cancer to the metabolism drug phenformin. Cancer Cell 23, 143–158 (2013).
Makowski, L. & Hayes, D. N. Role of LKB1 in lung cancer development. Br. J. Cancer 99, 683–688 (2008).
Morris, L. G. et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat. Genet. 45, 253–261 (2013).
Mogi, A. & Kuwano, H. TP53 mutations in nonsmall cell lung cancer. J. Biomed. Biotechnol. 2011, 583929 (2011).
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013).
Zeiler, M.D. & Fergus, R. Visualizing and understanding convolutional networks. In European Conference on Computer Vision. 818–833 (Springer International Publishing, New York, 2015).
Maaten, L. J. Pd Accelerating t-SNE using tree-based algorithms. J. Mach. Learn. Res. 15, 3221–3245 (2014).
Bonner, R. F. et al. Laser capture microdissection: molecular analysis of tissue. Science 278, 1481–1483 (1997). 1483.
Ninomiya, H. et al. Correlation between morphology and EGFR mutations in lung adenocarcinomas significance of the micropapillary pattern and the hobnail cell type. Lung Cancer 63, 235–240 (2009).
Warth, A. et al. EGFR, KRAS, BRAF and ALK gene alterations in lung adenocarcinomas: patient outcome, interplay with morphology and immunophenotype. Eur. Respir. J. 43, 872–883 (2014).
Sequist, L. V. et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 3, 75ra26 (2011).
Chiang, S. et al. IDH2 mutations define a unique subtype of breast cancer with altered nuclear polarity. Cancer Res. 76, 7118–7129 (2016).
Baas, A. F., Smit, L. & Clevers, H. LKB1 tumor suppressor protein: partaker in cell polarity. Trends Cell Biol. 14, 312–319 (2004).
Gloushankova, N., Ossovskaya, V., Vasiliev, J., Chumakov, P. & Kopnin, B. Changes in p53 expression can modify cell shape of ras-transformed fibroblasts and epitheliocytes. Oncogene 15, 2985–2989 (1997).
Yatabe, Y. et al. EGFR mutation is specific for terminal respiratory unit type adenocarcinoma. Am. J. Surg. Pathol. 29, 633–639 (2005).
Yoshida, A. et al. Comprehensive histologic analysis of ALK-rearranged lung carcinomas. Am. J. Surg. Pathol. 35, 1226–1234 (2011).
Rodig, S. J. et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin. Cancer Res. 15, 5216–5223 (2009).
Dearden, S., Stevens, J., Wu, Y.-L. & Blowers, D. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann. Oncol 24, 2371–2376 (2013).
Yu, J. et al. Mutation-specific antibodies for the detection of EGFR mutations in non-small-cell lung cancer. Clin. Cancer Res. 15, 3023–3028 (2009).
Houang, M. et al. EGFR mutation specific immunohistochemistry is a useful adjunct which helps to identify false negative mutation testing in lung cancer. Pathology 46, 501–508 (2014).
Dimou, A. et al. Standardization of epidermal growth factor receptor (EGFR) measurement by quantitative immunofluorescence and impact on antibody-based mutation detection in non–small cell lung cancer. Am. J. Pathol. 179, 580–589 (2011).
Schaumberg, A. J., Rubin, M. A. & Fuchs, T. J. H&E-stainedwhole slide deep learning predicts spop mutation state in prostate cancer. Preprint at https://doi.org/10.1101/064279 (2016).
Donovan, M. J. et al. A systems pathology model for predicting overall survival in patients with refractory, advanced non-small-cell lung cancer treated with gefitinib. Eur. J. Cancer 45, 1518–1526 (2009).
Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012).
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Hershey, S. et al. CNN architectures for large-scale audio classification. In 2017 IEEE International Conference on Acoustics, Speech and Signal Processing, 131–135 (New Orleans, LA, USA, 2017).
Hanley, J. A. & McNeil, B. J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143, 29–36 (1982).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Efron, B. & Tibshirani, R. J. An Introduction to the Bootstrap 56 (CRC Press, Boca Raton, FL, USA, 1994).
Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 20, 37–46 (1960).
McHugh, M. L. Interrater reliability: the kappa statistic. Biochem. Med. (Zagreb) 22, 276–282 (2012).
Acknowledgements
We would like to thank the Applied Bioinformatics Laboratories (ABL) at the NYU School of Medicine for providing bioinformatics support and helping with the analysis and interpretation of the data. The Applied Bioinformatics Laboratories are a Shared Resource, partially supported by the Cancer Center Support Grant, P30CA016087 (A.T.), at the Laura and Isaac Perlmutter Cancer Center (A.T.). For this work, we used computing resources at the High-Performance Computing Facility (HPC) at NYU Langone Medical Center. The slide images and the corresponding cancer information were uploaded from the Genomic Data Commons portal (https://gdc-portal.nci.nih.gov) and are in whole or in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/). These data were publicly available without restriction, authentication or authorization necessary. We thank the GDC help desk for providing assistance and information regarding the TCGA dataset. For the independent cohorts, we only used whole-slide images; the NYU dataset we used consists of slide images without identifiable information and therefore does not require approval according to both federal regulations and the NYU School of Medicine Institutional Review Board. For this same reason, written informed consent was not necessary. We thank C. Dickerson, from the Center for Biospecimen Research and Development (CBRD), for scanning the whole-slide images from the NYU Langone Medical Center. We also thank T. Papagiannakopoulos, H. Pass and K.-K. Wong or their valuable and constructive suggestions.
Author information
Authors and Affiliations
Contributions
N.C. performed the experiments; N.C., A.T. and N.R. designed the experiments; N.C. and T.S. wrote the code to achieve different tasks; T.S. gathered the mutation information and contributed to their analysis; M.S. helped identify cases validated by next-generation sequencing; A.L.M. and P.S.O. collected and labeled the independent cohorts. A.L.M, P.S.O and N.N. manually labeled the TCGA dataset; N.C., A.L.M., P.S.O., N.R. and A.T. contributed to the analysis of the data; D.F., N.R. and A.T. conceived and directed the project; N.C., A.T., N.R., A.L.M. and P.S.O. wrote the manuscript with the assistance and feedback of all the other co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Tables 1–7.
Rights and permissions
About this article
Cite this article
Coudray, N., Ocampo, P.S., Sakellaropoulos, T. et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med 24, 1559–1567 (2018). https://doi.org/10.1038/s41591-018-0177-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0177-5
This article is cited by
-
Deep learning in cancer genomics and histopathology
Genome Medicine (2024)
-
Pathogenomics for accurate diagnosis, treatment, prognosis of oncology: a cutting edge overview
Journal of Translational Medicine (2024)
-
A systematic analysis of deep learning in genomics and histopathology for precision oncology
BMC Medical Genomics (2024)
-
Oral epithelial dysplasia detection and grading in oral leukoplakia using deep learning
BMC Oral Health (2024)
-
Slideflow: deep learning for digital histopathology with real-time whole-slide visualization
BMC Bioinformatics (2024)