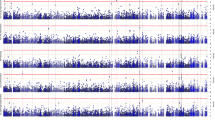
Abstract
The chances to develop Alzheimer’s disease (AD) result from a combination of genetic and non-genetic risk factors1, the latter likely being mediated by epigenetic mechanisms2. In the past, genome-wide association studies (GWAS) have identified an important number of risk loci associated with AD pathology3, but a causal relationship remains difficult to establish. In contrast, locus-specific or epigenome-wide association studies (EWAS) have revealed site-specific epigenetic alterations, which provide mechanistic insights for a particular risk gene but often lack the statistical power of GWAS4. Here, combining both approaches, we report a previously unidentified association of the peptidase M20-domain-containing protein 1 (PM20D1) with AD. We find that PM20D1 is a methylation and expression quantitative trait locus coupled to an AD-risk associated haplotype, which displays enhancer-like characteristics and contacts the PM20D1 promoter via a haplotype-dependent, CCCTC-binding-factor-mediated chromatin loop. Furthermore, PM20D1 is increased following AD-related neurotoxic insults at symptomatic stages in the APP/PS1 mouse model of AD and in human patients with AD who are carriers of the non-risk haplotype. In line, genetically increasing or decreasing the expression of PM20D1 reduces and aggravates AD-related pathologies, respectively. These findings suggest that in a particular genetic background, PM20D1 contributes to neuroprotection against AD.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Holtzman, D. M., Morris, J. C. & Goate, A. M. Alzheimer’s disease: the challenge of the second century. Sci. Transl. Med. 3, 77sr1 (2011).
Chouliaras, L. et al. Epigenetic regulation in the pathophysiology of Alzheimer’s disease. Prog. Neurobiol. 90, 498–510 (2010).
Scheltens, P. et al. Alzheimer’s disease. Lancet 388, 505–517 (2016).
Rakyan, V. K., Down, T. A., Balding, D. J. & Beck, S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 12, 529–541 (2011).
Sanchez-Mut, J. V. & Gräff, J. Epigenetic alterations in Alzheimer’s disease. Front. Behav. Neurosci. 9, 347 (2015).
Lunnon, K. et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat. Neurosci. 17, 1164–1170 (2014).
De Jager, P. L. et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat. Neurosci. 17, 1156–1163 (2014).
Siegmund, K. D. et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One 2, e895 (2007).
Sanchez-Mut, J. V. et al. DNA methylation map of mouse and human brain identifies target genes in Alzheimer’s disease. Brain 136, 3018–3027 (2013).
Gräff, J. et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature 483, 222–226 (2012).
D’Addario, C. et al. Transcriptional and epigenetic phenomena in peripheral blood cells of monozygotic twins discordant for Alzheimer’s disease, a case report. J. Neurol. Sci. 372, 211–216 (2017).
Hannon, E. et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat. Neurosci. 19, 48–54 (2016).
Stewart, S. E. et al. Genome-wide association study of obsessive–compulsive disorder. Mol. Psychiatry 18, 788–798 (2013).
Gamazon, E. R. et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol. Psychiatry 18, 340–346 (2013).
Sanchez-Mut, J. V. et al. Promoter hypermethylation of the phosphatase DUSP22 mediates PKA-dependent TAU phosphorylation and CREB activation in Alzheimer’s disease. Hippocampus 24, 363–368 (2014).
Sanchez-Mut, J. V. et al. Human DNA methylomes of neurodegenerative diseases show common epigenomic patterns. Transl. Psychiatry 6, e718 (2016).
Heyn, H. et al. DNA methylation contributes to natural human variation. Genome Res. 23, 1363–1372 (2013).
1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Li, H., Teo, Y. Y. & Tan, E. K. Patterns of linkage disequilibrium at PARK16 may explain variances in genetic-association studies. Mov. Disord. 30, 1335–1342 (2015).
Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010).
GTEx Consortium. Human genomics. The Genotype–Tissue Expression (GTEx) pilot analysis: multi-tissue gene regulation in humans. Science 348, 648–660 (2015).
Bulger, M. & Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–339 (2011).
Phillips, J. E. & Corces, V. G. CTCF: master weaver of the genome. Cell 137, 1194–1211 (2009).
Suzuki, M., Yamada, T., Kihara-Negishi, F., Sakurai, T. & Oikawa, T. Direct association between PU.1 and MeCP2 that recruits mSin3A–HDAC complex for PU.1-mediated transcriptional repression. Oncogene 22, 8688–8698 (2003).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Ashoor, H., Kleftogiannis, D., Radovanovic, A. & Bajic, V. B. DENdb: database of integrated human enhancers. Database (Oxford) 2015, bav085 (2015).
Smith, E. & Shilatifard, A. Enhancer biology and enhanceropathies. Nat. Struct. Mol. Biol. 21, 210–219 (2014).
Creyghton, M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931–21936 (2010).
Borchelt, D. R. et al. Accelerated amyloid deposition in the brains of transgenic mice coexpressing mutant presenilin 1 and amyloid precursor proteins. Neuron 19, 939–945 (1997).
Jankowsky, J. L. et al. Mutant presenilins specifically elevate the levels of the 42 residue β-amyloid peptide in vivo: evidence for augmentation of a 42-specific γ-secretase. Hum. Mol. Genet. 13, 159–170 (2004).
Long, J. Z. et al. The secreted enzyme PM20D1 regulates lipidated amino acid uncouplers of mitochondria. Cell 166, 424–435 (2016).
Feinberg, A. P. et al. Personalized epigenomic signatures that are stable over time and co-vary with body mass index. Sci. Transl. Med. 2, 49ra67 (2010).
Larrick, J. W., Larrick, J. W. & Mendelsohn, A. R. Uncoupling mitochondrial respiration for diabesity. Rejuvenation Res. 19, 337–340 (2016).
Kivipelto, M. & Mangialasche, F. Alzheimer’s disease: to what extent can Alzheimer’s disease be prevented? Nat. Rev. Neurol. 10, 552–553 (2014).
Maltby, V. E. et al. Differential methylation at MHC in CD4+ T cells is associated with multiple sclerosis independently of HLA-DRB1. Clin. Epigenetics 9, 71 (2017).
Simón-Sánchez, J. et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 41, 1308–1312 (2009).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-seq applications. Bioinformatics 27, 1571–1572 (2011).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Li, H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics 27, 718–719 (2011).
Watson, C. T. et al. Genome-wide DNA methylation profiling in the superior temporal gyrus reveals epigenetic signatures associated with Alzheimer’s disease. Genome Med. 8, 5 (2016).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Chen, M. H. & Yang, Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics 26, 580–581 (2010).
Guedj, M., Wojcik, J., Della-Chiesa, E., Nuel, G. & Forner, K. A fast, unbiased and exact allelic test for case–control association studies. Hum. Hered. 61, 210–221 (2006).
Braak, H. & Braak, E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991).
Heyn, H. et al. Distinct DNA methylomes of newborns and centenarians. Proc. Natl. Acad. Sci. USA 109, 10522–10527 (2012).
Klein, W. L. Aβ toxicity in Alzheimer’s disease: globular oligomers (ADDLs) as new vaccine and drug targets. Neurochem. Int. 41, 345–352 (2002).
Bibikova, M. et al. Genome-wide DNA methylation profiling using Infinium assay. Epigenomics 1, 177–200 (2009).
Fernandez, A. F. et al. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res. 19, 438–451 (2009).
Ruijter, J. M. et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37, e45 (2009).
Narayanan, M. et al. Common dysregulation network in the human prefrontal cortex underlies two neurodegenerative diseases. Mol. Syst. Biol. 10, 743 (2014).
Webster, J. A. et al. Genetic control of human brain transcript expression in Alzheimer’s disease. Am. J. Hum. Genet. 84, 445–458 (2009).
Hokama, M. et al. Altered expression of diabetes-related genes in Alzheimer’s disease brains: the Hisayama study. Cereb. Cortex 24, 2476–2488 (2014).
Court, F. et al. Long-range chromatin interactions at the mouse Igf2–H19 locus reveal a novel paternally expressed long noncoding RNA. Nucleic Acids Res 39, 5893–5906 (2011).
Acknowledgements
The work in the laboratory of J.G. is supported by the SYNAPSIS Foundation, the Béatrice Ederer-Weber Stiftung, the Floshield Foundation and the Alzheimer’s Association (grant no. NIRG-15-363964). The laboratory of D.M. is supported by the Foundation Jérôme Lejeune, Spanish Ministerio de Educación y Competitividad (grant no. BFU2014-53093). The laboratory of J.P.-T. is supported by the Spanish Ministerio de Economía, Industria y Competitividad and the FEDER programme from the EU (grant no. SAF2014-59469-R) and the CIBERNED. J.V.S.-M. is supported by a SYNAPSIS Foundation Fellowship for Advanced PostDocs and the Heidi Seiler-Stiftung foundation. H.H. is a Miguel Servet (CP14/00229) researcher funded by the Spanish Institute of Health Carlos III (ISCIII). B.A.S. is an EMBO long-term fellow (ALTF 1605-2014, Marie Curie Actions, LTFCOFUND2013, GA-2013-609409). A.M.-S. is a recipient of a FPI PhD studentship from MINECO. M.E. is an ICREA Research Professor. J.G. is an MQ fellow and a NARSAD Independent Investigator.
Author information
Authors and Affiliations
Contributions
J.V.S.-M., H.H., M.E. and J.G. conceived the project and designed the experiments; J.V.S.-M., B.A.S., L.D., P.G.-E., L.G., A.M.-S. and D.M. performed the experiments; J.V.S.-M., E.V. and S.S. performed the bioinformatics analysis of the data; J.P.-T., I.F., B.S., D.M. and M.E. contributed to the interpretation of the results; and J.V.S.-M. and J.G. wrote the paper, with input and comments by all of the authors.
Corresponding author
Ethics declarations
Competing interests
A provisional patent application has been filed on the use of PM20D1 methylation and haplotype as biomarkers for Alzheimer’s disease (International Application Number PCT/EP2017/067848), with J.V.S.-M., H.H., M.E. and J.G. listed as inventors.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Tables and Figures
Supplementary Tables 1–3 and Supplementary Figures 1–7
Rights and permissions
About this article
Cite this article
Sanchez-Mut, J.V., Heyn, H., Silva, B.A. et al. PM20D1 is a quantitative trait locus associated with Alzheimer’s disease. Nat Med 24, 598–603 (2018). https://doi.org/10.1038/s41591-018-0013-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0013-y
This article is cited by
-
Serum PM20D1 levels in patients with idiopathic pulmonary arterial hypertension and its clinical significance
BMC Cardiovascular Disorders (2024)
-
Systemic interindividual epigenetic variation in humans is associated with transposable elements and under strong genetic control
Genome Biology (2023)
-
Neurofilament-light chain quantification by Simoa and Ella in plasma from patients with dementia: a comparative study
Scientific Reports (2023)
-
Circulating cell-free DNA methylation mirrors alterations in cerebral patterns in epilepsy
Clinical Epigenetics (2022)
-
Distinct sex-specific DNA methylation differences in Alzheimer’s disease
Alzheimer's Research & Therapy (2022)