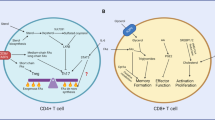
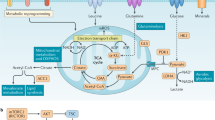
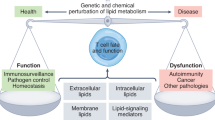
Abstract
This Review explores the interplay between T cell activation and cell metabolism and highlights how metabolites serve two pivotal functions in shaping the immune response. Traditionally, T cell activation has been characterized by T cell antigen receptor–major histocompatibility complex interaction (signal 1), co-stimulation (signal 2) and cytokine signaling (signal 3). However, recent research has unveiled the critical role of metabolites in this process. Firstly, metabolites act as signal propagators that aid in the transmission of core activation signals, such as specific lipid species that are crucial at the immune synapse. Secondly, metabolites also function as unique signals that influence immune differentiation pathways, such as amino acid-induced mTORC1 signaling. Metabolites also play a substantial role in epigenetic remodeling, by directly modifying histones, altering gene expression and influencing T cell behavior. This Review discusses how T cells integrate nutrient sensing with activating stimuli to shape their differentiation and sensitivity to metabolites. We underscore the integration of immunological and metabolic inputs in T cell function and suggest that metabolite availability is a fundamental determinant of adaptive immune responses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Smith-Garvin, J. E., Koretzky, G. A. & Jordan, M. S. T cell activation. Annu. Rev. Immunol. 27, 591–619 (2009).
Pathan-Chhatbar, S. et al. Direct regulation of the T cell antigen receptor’s activity by cholesterol. Front. Cell Dev. Biol. 8, 615996 (2021).
Wu, W., Shi, X. & Xu, C. Regulation of T cell signalling by membrane lipids. Nat. Rev. Immunol. 16, 690–701 (2016).
He, H. T., Lellouch, A. & Marguet, D. Lipid rafts and the initiation of T cell receptor signaling. Semin. Immunol. 17, 23–33 (2005).
Jury, E. C., Flores-Borja, F. & Kabouridis, P. S. Lipid rafts in T cell signalling and disease. Semin. Cell Dev. Biol. 18, 608–615 (2007).
Kabouridis, P. S. Lipid rafts in T cell receptor signalling. Mol. Membr. Biol. 23, 49–57 (2006).
Chen, Y. et al. Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility. Mol. Cell 82, 1278–1287 (2022).
Bensinger, S. J. et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell 134, 97–111 (2008).
Wilfahrt, D. et al. Histone deacetylase 3 represses cholesterol efflux during CD4+ T-cell activation. Elife 10, e70978 (2021).
Lu, W. et al. The phosphatidylinositol-transfer protein Nir3 promotes PI(4,5)P2 replenishment in response to TCR signaling during T cell development and survival. Nat. Immunol. 24, 136–147 (2022).
Edwards-Hicks, J. et al. Phosphoinositide acyl chain saturation drives CD8+ effector T cell signaling and function. Nat. Immunol. 24, 516–530 (2023).
Kremer, K. N. et al. LPA suppresses T cell function by altering the cytoskeleton and disrupting immune synapse formation. Proc. Natl Acad. Sci. USA 119, e2118816119 (2022).
Mathew, D. et al. LPA 5 is an inhibitory receptor that suppresses CD8 T-cell cytotoxic function via disruption of early TCR signaling. Front Immunol. 10, 1159 (2019).
Zhang, M. S. et al. Glycerol monolaurate (GML) induces filopodia formation by disrupting the association between LAT and SLP-76 microclusters. Sci. Signal 11, eaan9095 (2018).
Castellano, F. & Molinier-Frenkel, V. Control of T-cell activation and signaling by amino-acid catabolizing enzymes. Front. Cell Dev. Biol. 8, 613416 (2020).
Wu, J. et al. Asparagine enhances LCK signalling to potentiate CD8+ T-cell activation and anti-tumour responses. Nat. Cell Biol. 23, 75–86 (2021).
Hope, H. C. et al. Coordination of asparagine uptake and asparagine synthetase expression modulates CD8+ T cell activation. JCI Insight 6, e137761 (2021).
Geiger, R. et al. L-Arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell 167, 829–842 (2016).
Carr, E. L. et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 185, 1037–1044 (2010).
Wang, R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011).
Johnson, M. O. et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell 175, 1780–1795 (2018).
Ginefra, P., Hope, H. C., Spagna, M., Zecchillo, A. & Vannini, N. Ionic regulation of T-cell function and anti-tumour immunity. Int. J. Mol. Sci. 22, 13668 (2021).
Lötscher, J. et al. Magnesium sensing via LFA-1 regulates CD8+ T cell effector function. Cell 185, 585–602 (2022).
Luik, R. M., Wang, B., Prakriya, M., Wu, M. M. & Lewis, R. S. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature 454, 538–542 (2008).
Hogan, P. G. Calcium–NFAT transcriptional signalling in T cell activation and T cell exhaustion. Cell Calcium 63, 66–69 (2017).
Vaeth, M. et al. Store-operated Ca2+ entry controls clonal expansion of T cells through metabolic reprogramming. Immunity 47, 664–679 (2017).
Wang, Y. H., Tao, A. Y., Vaeth, M. & Feske, S. Calcium regulation of T cell metabolism. Curr. Opin. Physiol. 17, 207–223 (2020).
Balmer, M. L. et al. Memory CD8+ T cells balance pro- and anti-inflammatory activity by reprogramming cellular acetate handling at sites of infection. Cell Metab. 32, 457–467 (2020).
Balmer, M. L. et al. Memory CD8+ T cells require increased concentrations of acetate induced by stress for optimal function. Immunity 44, 1312–1324 (2016).
Desdín-Micó, G., Soto-Heredero, G. & Mittelbrunn, M. Mitochondrial activity in T cells. Mitochondrion 41, 51–57 (2018).
Lisci, M. & Griffiths, G. M. Arming a killer: mitochondrial regulation of CD8+ T cell cytotoxicity. Trends Cell Biol. 33, 138–147 (2023).
Quintana, A. et al. T cell activation requires mitochondrial translocation to the immunological synapse. Proc. Natl Acad. Sci. USA 104, 14418–14423 (2007).
Yoast, R. E. et al. The mitochondrial Ca2+ uniporter is a central regulator of interorganellar Ca2+ transfer and NFAT activation. J. Biol. Chem. 297, 101174 (2021).
Wu, H. et al. Genetic ablation of the mitochondrial calcium uniporter (MCU) does not impair T cell-mediated immunity in vivo. Front Pharmacol. 12, 734078 (2021).
Sena, L. A. et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236 (2013).
Häusler, D. et al. High dose vitamin D exacerbates central nervous system autoimmunity by raising T-cell excitatory calcium. Brain 142, 2737–2755 (2019).
Liang, Y. et al. Retinoic acid modulates hyperactive T cell responses and protects vitamin A-deficient mice against persistent lymphocytic choriomeningitis virus infection. J. Immunol. 204, 2984–2994 (2020).
Ho, P. C. et al. Phosphoenolpyruvate Is a metabolic checkpoint of anti-tumor T cell responses. Cell 162, 1217–1228 (2015).
Bunse, L. et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat. Med. 24, 1192–1203 (2018).
Boomer, J. S. & Green, J. M. An enigmatic tail of CD28 signaling. Cold Spring Harb. Perspect. Biol. 2, a002436 (2010).
Chapman, N. M., Boothby, M. R. & Chi, H. Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20, 55–70 (2020).
Benson, M. J., Pino-Lagos, K., Rosemblatt, M. & Noelle, R. J. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J. Exp. Med. 204, 1765–1774 (2007).
Basu, R. et al. IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the TH17 cell-iTreg cell balance. Nat. Immunol. 16, 286–295 (2015).
Friesen, L. R., Gu, B. & Kim, C. H. A ligand-independent fast function of RARα promotes exit from metabolic quiescence upon T cell activation and controls T cell differentiation. Mucosal Immunol. 14, 100–112 (2020).
Xu, K. et al. Glycolysis fuels phosphoinositide 3-kinase signaling to bolster T cell immunity. Science 371, 405–410 (2021).
Xu, K. et al. Glycolytic ATP fuels phosphoinositide 3-kinase signaling to support effector T helper 17 cell responses. Immunity 54, 976–987 (2021).
Jacobs, S. R. et al. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J. Immunol. 180, 4476–4486 (2008).
Zhu, M., Foreman, D. P., O’Brien, S. A., Jin, Y. & Zhang, W. Phospholipase D in TCR-mediated signaling and T cell activation. J. Immunol. 200, 2165–2173 (2018).
Scheffel, M. J. et al. N-acetyl cysteine protects anti-melanoma cytotoxic T cells from exhaustion induced by rapid expansion via the downmodulation of Foxo1 in an Akt-dependent manner. Cancer Immunol. Immunother. 67, 691–702 (2018).
Sengupta, S., Peterson, T. R. & Sabatini, D. M. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40, 310–322 (2010).
Szwed, A., Kim, E. & Jacinto, E. Regulation and metabolic functions of mTORC1 and mTORC2. Physiol. Rev. 101, 1371–1426 (2021).
Zeng, H. et al. mTORC1 couples immune signals and metabolic programming to establish Treg-cell function. Nature 499, 485–490 (2013).
Zeng, H. et al. mTORC1 and mTORC2 kinase signaling and glucose metabolism drive follicular helper T cell differentiation. Immunity 45, 540–554 (2016).
Finlay, D. K. et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J. Exp. Med. 209, 2441–2453 (2012).
Shi, L. Z. et al. HIF1α–dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208, 1367–1376 (2011).
Tan, H. et al. Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity 46, 488–503 (2017).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Sinclair, L. V. et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 14, 500–508 (2013).
Nakaya, M. et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity 40, 692–705 (2014).
Eil, R. et al. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature 537, 539–543 (2016).
Yin, S., Liu, L. & Gan, W. The roles of post-translational modifications on mTOR signaling. Int. J. Mol. Sci. 22, 1784 (2021).
Su, W. et al. Protein prenylation drives discrete signaling programs for the differentiation and maintenance of effector Treg cells. Cell Metab. 32, 996–1011 (2020).
Yang, W. et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat. Commun. 11, 4457 (2020).
Fu, G. et al. Metabolic control of Tfh cells and humoral immunity by phosphatidylethanolamine. Nature 595, 724–729 (2021).
Dong, C. Cytokine regulation and function in T cells. Annu. Rev. Immunol. 39, 51–76 (2021).
Firestein, G. S. Evolving concepts of rheumatoid arthritis. Nature 423, 356–361 (2003).
Walker, L. S. K. & von Herrath, M. CD4 T cell differentiation in type 1 diabetes. Clin. Exp. Immunol. 183, 16–29 (2016).
Ross, S. H. & Cantrell, D. A. Signaling and function of interleukin-2 in T lymphocytes. Annu. Rev. Immunol. 36, 411–433 (2018).
Chisolm, D. A. et al. CCCTC-binding factor translates interleukin 2- and α-ketoglutarate-sensitive metabolic changes in T cells into context-dependent gene programs. Immunity 47, 251–267 (2017).
Chang, C. H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013).
Wang, Z. et al. Iron drives T helper cell pathogenicity by promoting RNA-binding protein PCBP1-mediated proinflammatory cytokine production. Immunity 49, 80–92 (2018).
Villarino, A. V et al. A central role for STAT5 in the transcriptional programing of T helper cell metabolism. Sci. Immunol. 7, eabl9467 (2022).
Ray, J. P. et al. The interleukin-2–mTORc1 kinase axis defines the signaling, differentiation, and metabolism of T helper 1 and follicular B helper T cells. Immunity 43, 690–702 (2015).
Mo, F. et al. An engineered IL-2 partial agonist promotes CD8+ T cell stemness. Nature 597, 544–548 (2021).
He, Y. et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8+ T cell immunity. Cell Metab. 33, 988–1000 (2021).
Wynn, T. A. Type 2 cytokines: mechanisms and therapeutic strategies. Nat. Rev. Immunol. 15, 271–282 (2015).
Morgan, R. et al. N-acetylglucosaminyltransferase V (Mgat5)-mediated N-glycosylation negatively regulates Th1 cytokine production by T cells. J. Immunol. 173, 7200–7208 (2004).
Jiang, H. et al. LZTFL1 upregulated by all-trans retinoic acid during CD4+ T cell activation enhances IL-5 production. J. Immunol. 196, 1081–1090 (2016).
Nalleweg, N. et al. IL-9 and its receptor are predominantly involved in the pathogenesis of UC. Gut 64, 743–755 (2015).
Wilhelm, C. et al. An IL-9 fate reporter demonstrates the induction of an innate IL-9 response in lung inflammation. Nat. Immunol. 12, 1071–1077 (2011).
Roy, S. et al. EGFR-HIF1α signaling positively regulates the differentiation of IL-9 producing T helper cells. Nat. Commun. 12, 3182 (2021).
Schwartz, D. M. et al. Retinoic acid receptor alpha represses a Th9 transcriptional and epigenomic program to reduce allergic pathology. Immunity 50, 106–120 (2019).
Asadzadeh, Z. et al. The paradox of Th17 cell functions in tumor immunity. Cell Immunol. 322, 15–25 (2017).
Tan, S. et al. The pro-inflammatory effect of triglyceride on human CD4+ T cells and experimental autoimmune uveitis. Clin. Immunol. 240, 109056 (2022).
Paik, D. et al. Human gut bacteria produce ΤΗ17-modulating bile acid metabolites. Nature 603, 907–912 (2022).
Duan, J. et al. Endoplasmic reticulum stress in the intestinal epithelium initiates purine metabolite synthesis and promotes Th17 cell differentiation in the gut. Immunity 56, 1115–1131 (2023).
Berod, L. et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 20, 1327–1333 (2014).
Wang, C. et al. CD5L/AIM regulates lipid biosynthesis and restrains Th17 cell pathogenicity. Cell 163, 1413–1427 (2015).
Wagner, A. et al. Metabolic modeling of single Th17 cells reveals regulators of autoimmunity. Cell 184, 4168–4185 (2021).
Lian, G. et al. Glutathione de novo synthesis but not recycling process coordinates with glutamine catabolism to control redox homeostasis and directs murine T cell differentiation. Elife 7, e36158 (2018).
Gerriets, V. A. et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 125, 194–207 (2015).
Zenewicz, L. A. et al. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity 29, 947–957 (2008).
Ouyang, W. & O’Garra, A. IL-10 family cytokines IL-10 and IL-22: from basic science to clinical translation. Immunity 50, 871–891 (2019).
Ford, B. R. et al. Tumor microenvironmental signals reshape chromatin landscapes to limit the functional potential of exhausted T cells. Sci. Immunol. 7, eabj9123 (2022).
Quezada, L. K. et al. Early transcriptional and epigenetic divergence of CD8+ T cells responding to acute versus chronic infection. PLoS Biol. 21, e3001983 (2023).
Byvoet, P., Shepherd, G. R., Hardin, J. M. & Noland, B. J. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch. Biochem. Biophys. 148, 558–567 (1972).
Murray, K. The occurrence of epsilon-N-methyl lysine in histones. Biochemistry 3, 10–15 (1964).
Fischle, W., Franz, H., Jacobs, S. A., Allis, C. D. & Khorasanizadeh, S. Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. J. Biol. Chem. 283, 19626–19635 (2008).
Greer, E. L. & Shi, Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 (2012).
Bernstein, B. E. et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl Acad. Sci. USA 99, 8695–8700 (2002).
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Correa, L. O., Jordan, M. S. & Carty, S. A. DNA methylation in T-cell development and differentiation. Crit. Rev. Immunol. 40, 135–156 (2020).
Lyko, F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92 (2017).
Zhao, T. & Lum, J. J. Methionine cycle-dependent regulation of T cells in cancer immunity. Front Oncol. 12, 969563 (2022).
Roy, D. G. et al. Methionine metabolism shapes T helper cell responses through regulation of epigenetic reprogramming. Cell Metab. 31, 250–266 (2020).
Bian, Y. et al. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature 585, 277–282 (2020).
DeVorkin, L. et al. Autophagy regulation of metabolism is required for CD8+ T cell anti-tumor immunity. Cell Rep. 27, 502–513 (2019).
Scourzic, L., Mouly, E. & Bernard, O. A. TET proteins and the control of cytosine demethylation in cancer. Genome Med. 7, 9 (2015).
Klysz, D. et al. Glutamine-dependent α-ketoglutarate production regulates the balance between T helper 1 cell and regulatory T cell generation. Sci. Signal. 8, ra97 (2015).
Matias, M. I. et al. Regulatory T cell differentiation is controlled by αKG-induced alterations in mitochondrial metabolism and lipid homeostasis. Cell Rep. 37, 109911 (2021).
Yang, M. & Pollard, P. J. Succinate: a new epigenetic hacker. Cancer Cell 23, 709–711 (2013).
Chen, X. et al. Succinate dehydrogenase/complex II is critical for metabolic and epigenetic regulation of T cell proliferation and inflammation. Sci. Immunol. 7, eabm8161 (2022).
Tyrakis, P. A. et al. The immunometabolite S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 540, 236–241 (2016).
Marmorstein, R. & Zhou, M. M. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 6, a018762 (2014).
Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Yang, X. J. & Seto, E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26, 5310–5318 (2007).
Shahbazian, M. D. & Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 (2007).
Lee, K. K. & Workman, J. L. Histone acetyltransferase complexes: one size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295 (2007).
Schmitt, D. L. & An, S. Spatial organization of metabolic enzyme complexes in cells. Biochemistry 56, 3184–3196 (2017).
Pietrocola, F., Galluzzi, L., Bravo-San Pedro, J. M., Madeo, F. & Kroemer, G. Acetyl coenzyme a: a central metabolite and second messenger. Cell Metab. 21, 805–821 (2015).
Wellen, K. E. et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009).
Peng, M. et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016).
Bailis, W. et al. Distinct modes of mitochondrial metabolism uncouple T cell differentiation and function. Nature 571, 403–407 (2019).
Nakajima, T. et al. ACC1-expressing pathogenic T helper 2 cell populations facilitate lung and skin inflammation in mice. J. Exp. Med. 218, e20210639 (2021).
Puleston, D. J. et al. Polyamine metabolism is a central determinant of helper T cell lineage fidelity. Cell 184, 4186–4202 (2021).
Hochrein, S. M. et al. The glucose transporter GLUT3 controls T helper 17 cell responses through glycolytic-epigenetic reprogramming. Cell Metab. 34, 516–532 (2022).
Qiu, J. et al. Acetate promotes T cell effector function during glucose restriction. Cell Rep. 27, 2063–2074 (2019).
Chowdhury, S. et al. Intracellular Acetyl CoA potentiates the therapeutic efficacy of antitumor CD8+ T cells. Cancer Res. 82, 2640–2655 (2022).
Vodnala, S. K. et al. T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 363, eaau0135 (2019).
Drummond, D. C. et al. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu. Rev. Pharmacol. Toxicol. 45, 495–528 (2005).
Feng, Q. et al. Lactate increases stemness of CD8+ T cells to augment anti-tumor immunity. Nat. Commun. 13, 4981 (2022).
Park, J. et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR–S6K pathway. Mucosal Immunol. 8, 80–93 (2014).
Kibbie, J. J. et al. Butyrate directly decreases human gut lamina propria CD4 T cell function through histone deacetylase (HDAC) inhibition and GPR43 signaling. Immunobiology 226, 152126 (2021).
Zhang, D. et al. Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 (2019).
Caslin, H. L., Abebayehu, D., Pinette, J. A. & Ryan, J. J. Lactate is a metabolic mediator that shapes immune cell fate and function. Front. Physiol. 12, 688485 (2021).
Jiang, G., Li, C., Lu, M., Lu, K. & Li, H. Protein lysine crotonylation: past, present, perspective. Cell Death Dis. 12, 703 (2021).
Kebede, A. F. et al. Histone propionylation is a mark of active chromatin. Nat. Struct. Mol. Biol. 24, 1048–1056 (2017).
Chen, Y. et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell Proteomics 6, 812–819 (2007).
Karthik Varanasi, S., Vijaya Kumar, S. & Rouse, B. T. Determinants of tissue-specific metabolic adaptation of T cells. Cell Metab. 32, 908–919 (2020).
Liu, X., Hoft, D. F. & Peng, G. Tumor microenvironment metabolites directing T cell differentiation and function. Trends Immunol. 43, 132–147 (2022).
Carrer, A. & Wellen, K. E. Metabolism and epigenetics: a link cancer cells exploit. Curr. Opin. Biotechnol. 34, 23–29 (2015).
Author information
Authors and Affiliations
Contributions
D.W. wrote the initial draft of the manuscript and designed the figures. G.M.D. supervised the writing of and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.W. declares no competing interests. G.M.D. has filed/awarded patents and founded companies around modulation of immune metabolism for disease treatment.
Peer review
Peer review information
Nature Immunology thanks Ping-Chih Ho and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Ioana Staicu, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wilfahrt, D., Delgoffe, G.M. Metabolic waypoints during T cell differentiation. Nat Immunol 25, 206–217 (2024). https://doi.org/10.1038/s41590-023-01733-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-023-01733-5