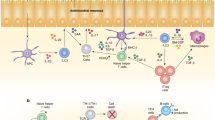
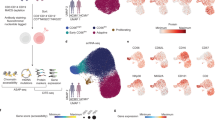
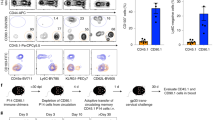
Abstract
The ability of vertebrates to ‘remember’ previous infections had once been attributed exclusively to adaptive immunity. We now appreciate that innate lymphocytes also possess memory properties akin to those of adaptive immune cells. In this Review, we draw parallels from T cell biology to explore the key features of immune memory in innate lymphocytes, including quantity, quality, and location. We discuss the signals that trigger clonal or clonal-like expansion in innate lymphocytes, and highlight recent studies that shed light on the complex cellular and molecular crosstalk between metabolism, epigenetics, and transcription responsible for differentiating innate lymphocyte responses towards a memory fate. Additionally, we explore emerging evidence that activated innate lymphocytes relocate and establish themselves in specific peripheral tissues during infection, which may facilitate an accelerated response program akin to those of tissue-resident memory T cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Riedel, S. Edward Jenner and the history of smallpox and vaccination. Proc. (Bayl. Univ. Med Cent.) 18, 21–25 (2005).
Boylston, A. The origins of inoculation. J. R. Soc. Med. 105, 309–313 (2012).
Sun, J. C., Beilke, J. N. & Lanier, L. L. Adaptive immune features of natural killer cells. Nature 457, 557–561 (2009).
Dokun, A. O. et al. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol. 2, 951–956 (2001).
Daniels, K. A. et al. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J. Exp. Med. 194, 29–44 (2001).
Brown, M. G. et al. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science 292, 934–937 (2001).
Arase, H., Mocarski, E. S., Campbell, A. E., Hill, A. B. & Lanier, L. L. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296, 1323–1326 (2002).
Grassmann, S. et al. Distinct surface expression of activating receptor Ly49H drives differential expansion of NK cell clones upon murine cytomegalovirus infection. Immunity 50, 1391–1400 (2019).
Reeves, R. K. et al. Antigen-specific NK cell memory in rhesus macaques. Nat. Immunol. 16, 927–932 (2015).
Schlums, H. et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42, 443–456 (2015).
Lopez-Verges, S. et al. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl Acad. Sci. USA 108, 14725–14732 (2011).
Lee, J. et al. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 42, 431–442 (2015).
Ty, M. et al. Malaria-driven expansion of adaptive-like functional CD56-negative NK cells correlates with clinical immunity to malaria. Sci. Transl. Med. 15, eadd9012 (2023).
Hart, G. T. et al. Adaptive NK cells in people exposed to Plasmodium falciparum correlate with protection from malaria. J. Exp. Med. 216, 1280–1290 (2019).
Ruckert, T., Lareau, C. A., Mashreghi, M. F., Ludwig, L. S. & Romagnani, C. Clonal expansion and epigenetic inheritance of long-lasting NK cell memory. Nat. Immunol. 23, 1551–1563 (2022).
Tomasec, P. et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287, 1031 (2000).
Heatley, S. L. et al. Polymorphism in human cytomegalovirus UL40 impacts on recognition of human leukocyte antigen-E (HLA-E) by natural killer cells. J. Biol. Chem. 288, 8679–8690 (2013).
Hammer, Q. et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol. 19, 453–463 (2018).
Ulbrecht, M. et al. Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J. Immunol. 164, 5019–5022 (2000).
Netea, M. G. et al. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 20, 375–388 (2020).
Naik, S. et al. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 550, 475–480 (2017).
Chen, L. & Flies, D. B. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 13, 227–242 (2013).
Nabekura, T. et al. Costimulatory molecule DNAM-1 is essential for optimal differentiation of memory natural killer cells during mouse cytomegalovirus infection. Immunity 40, 225–234 (2014).
Nabekura, T. et al. Cutting edge: NKG2D signaling enhances NK cell responses but alone is insufficient to drive expansion during mouse cytomegalovirus infection. J. Immunol. 199, 1567–1571 (2017).
Nabekura, T. & Lanier, L. L. Activating receptors for self-MHC class I enhance effector functions and memory differentiation of NK cells during mouse cytomegalovirus infection. Immunity 45, 74–82 (2016).
Liu, L. L. et al. Critical role of CD2 co-stimulation in adaptive natural killer cell responses revealed in NKG2C-deficient humans. Cell Rep. 15, 1088–1099 (2016).
Krug, A. et al. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity 21, 107–119 (2004).
Abbas, A. et al. The activation trajectory of plasmacytoid dendritic cells in vivo during a viral infection. Nat. Immunol. 21, 983–997 (2020).
Valente, M. et al. Novel mouse models based on intersectional genetics to identify and characterize plasmacytoid dendritic cells. Nat. Immunol. 24, 714–728 (2023).
Lio, C. W. et al. cGAS–STING signaling regulates initial innate control of cytomegalovirus infection. J. Virol. 90, 7789–7797 (2016).
Paijo, J. et al. cGAS senses human cytomegalovirus and induces type I interferon responses in human monocyte-derived cells. PLoS Pathog. 12, e1005546 (2016).
Madera, S. et al. Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J. Exp. Med. 213, 225–233 (2016).
Weizman, O. E. et al. ILC1 confer early host protection at initial sites of viral infection. Cell 171, 795–808 (2017).
Rolle, A. et al. IL-12-producing monocytes and HLA-E control HCMV-driven NKG2C+ NK cell expansion. J. Clin. Invest. 124, 5305–5316 (2014).
Sun, J. C. et al. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J. Exp. Med. 209, 947–954 (2012).
Andrews, D. M., Scalzo, A. A., Yokoyama, W. M., Smyth, M. J. & Degli-Esposti, M. A. Functional interactions between dendritic cells and NK cells during viral infection. Nat. Immunol. 4, 175–181 (2003).
Chaix, J. et al. Cutting edge: priming of NK cells by IL-18. J. Immunol. 181, 1627–1631 (2008).
Wiedemann, G. M. et al. Divergent role for STAT5 in the adaptive responses of natural killer cells. Cell Rep. 33, 108498 (2020).
Kaech, S. M. & Cui, W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012).
Wiedemann, G. M. et al. Deconvoluting global cytokine signaling networks in natural killer cells. Nat. Immunol. 22, 627–638 (2021).
Bekiaris, V. et al. Ly49H+ NK cells migrate to and protect splenic white pulp stroma from murine cytomegalovirus infection. J. Immunol. 180, 6768–6776 (2008).
Dokun, A. O., Chu, D. T., Yang, L., Bendelac, A. S. & Yokoyama, W. M. Analysis of in situ NK cell responses during viral infection. J. Immunol. 167, 5286–5293 (2001).
Andrews, D. M. et al. NK1.1+ cells and murine cytomegalovirus infection: what happens in situ? J. Immunol. 166, 1796–1802 (2001).
Salazar-Mather, T. P., Ishikawa, R. & Biron, C. A. NK cell trafficking and cytokine expression in splenic compartments after IFN induction and viral infection. J. Immunol. 157, 3054–3064 (1996).
Hsu, K. M., Pratt, J. R., Akers, W. J., Achilefu, S. I. & Yokoyama, W. M. Murine cytomegalovirus displays selective infection of cells within hours after systemic administration. J. Gen. Virol. 90, 33–43 (2009).
Flommersfeld, S. et al. Fate mapping of single NK cells identifies a type 1 innate lymphoid-like lineage that bridges innate and adaptive recognition of viral infection. Immunity 54, 2288–2304 (2021).
Diaz-Salazar, C. et al. Cell-intrinsic adrenergic signaling controls the adaptive NK cell response to viral infection. J. Exp. Med. 217, e20190549 (2020).
Benechet, A. P. et al. Dynamics and genomic landscape of CD8+ T cells undergoing hepatic priming. Nature 574, 200–205 (2019).
Kern, M. et al. Virally infected mouse liver endothelial cells trigger CD8+ T-cell immunity. Gastroenterology 138, 336–346 (2010).
Wang, Y. et al. Vaginal type-II mucosa is an inductive site for primary CD8+ T-cell mucosal immunity. Nat. Commun. 6, 6100 (2015).
Weizman, O. E. et al. Mouse cytomegalovirus-experienced ILC1s acquire a memory response dependent on the viral glycoprotein m12. Nat. Immunol. 20, 1004–1011 (2019).
Aguilar, O. A. et al. A viral immunoevasin controls innate immunity by targeting the prototypical natural killer cell receptor family. Cell 169, 58–71 (2017).
Martinez-Gonzalez, I. et al. Allergen-experienced group 2 innate lymphoid cells acquire memory-like properties and enhance allergic lung inflammation. Immunity 45, 198–208 (2016).
Verma, M. et al. The molecular and epigenetic mechanisms of innate lymphoid cell (ILC) memory and its relevance for asthma. J. Exp. Med. 218, e20201354 (2021).
Serafini, N. et al. Trained ILC3 responses promote intestinal defense. Science 375, 859–863 (2022).
Naik, S. & Fuchs, E. Inflammatory memory and tissue adaptation in sickness and in health. Nature 607, 249–255 (2022).
Halper-Stromberg, A. & Jabri, B. Maladaptive consequences of inflammatory events shape individual immune identity. Nat. Immunol. 23, 1675–1686 (2022).
Busch, D. H. & Pamer, E. G. T cell affinity maturation by selective expansion during infection. J. Exp. Med. 189, 701–710 (1999).
Adams, N. M. et al. Cytomegalovirus infection drives avidity selection of natural killer cells. Immunity 50, 1381–1390 (2019).
Chapman, N. M., Boothby, M. R. & Chi, H. Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20, 55–70 (2020).
Frauwirth, K. A. et al. The CD28 signaling pathway regulates glucose metabolism. Immunity 16, 769–777 (2002).
Ma, E. H. et al. Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8+ T cells. Immunity 51, 856–870 (2019).
Xu, K. et al. Glycolysis fuels phosphoinositide 3-kinase signaling to bolster T cell immunity. Science 371, 405–410 (2021).
Phan, A. T. et al. Constitutive glycolytic metabolism supports CD8+ T cell effector memory differentiation during viral infection. Immunity 45, 1024–1037 (2016).
Peng, M. et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016).
Sheppard, S. et al. Lactate dehydrogenase A-dependent aerobic glycolysis promotes natural killer cell anti-viral and anti-tumor function. Cell Rep. 35, 109210 (2021).
Loftus, R. M. et al. Amino acid-dependent cMyc expression is essential for NK cell metabolic and functional responses in mice. Nat. Commun. 9, 2341 (2018).
Liu, W. et al. Lactate regulates cell cycle by remodeling the anaphase promoting complex. Nature 616, 790–797 (2023).
Wang, R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011).
Littwitz-Salomon, E. et al. Metabolic requirements of NK cells during the acute response against retroviral infection. Nat. Commun. 12, 5376 (2021).
Dong, H. et al. The IRE1 endoplasmic reticulum stress sensor activates natural killer cell immunity in part by regulating c-Myc. Nat. Immunol. 20, 865–878 (2019).
Victorino, F. et al. HIF1α is required for NK cell metabolic adaptation during virus infection. eLife 10, e68484 (2021).
Ni, J. et al. Single-cell RNA sequencing of tumor-infiltrating NK cells reveals that inhibition of transcription factor HIF-1α unleashes NK cell activity. Immunity 52, 1075–1087 (2020).
Pearce, E. L. et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107 (2009).
Cui, G. et al. IL-7-induced glycerol transport and TAG synthesis promotes memory CD8+ T cell longevity. Cell 161, 750–761 (2015).
Raud, B. et al. Etomoxir actions on regulatory and memory T cells are independent of Cpt1a-mediated fatty acid oxidation. Cell Metab. 28, 504–515 (2018).
van der Windt, G. J. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012).
O’Sullivan, T. E., Johnson, L. R., Kang, H. H. & Sun, J. C. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity 43, 331–342 (2015).
Cichocki, F. et al. ARID5B regulates metabolic programming in human adaptive NK cells. J. Exp. Med. 215, 2379–2395 (2018).
Arts, R. J. et al. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 24, 807–819 (2016).
Bekkering, S. et al. Metabolic induction of trained immunity through the mevalonate pathway. Cell 172, 135–146 (2018).
Cheng, S. C. et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (2014).
Wellen, K. E. et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009).
Mentch, S. J. et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 22, 861–873 (2015).
Carey, B. W., Finley, L. W., Cross, J. R., Allis, C. D. & Thompson, C. B. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416 (2015).
Dai, Z., Ramesh, V. & Locasale, J. W. The evolving metabolic landscape of chromatin biology and epigenetics. Nat. Rev. Genet. 21, 737–753 (2020).
Yu, B. et al. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol. 18, 573–582 (2017).
Russ, B. E. et al. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8+ T cell differentiation. Immunity 41, 853–865 (2014).
Soriano-Baguet, L. & Brenner, D. Metabolism and epigenetics at the heart of T cell function. Trends Immunol. 44, 231–244 (2023).
Wenes, M. et al. The mitochondrial pyruvate carrier regulates memory T cell differentiation and antitumor function. Cell Metab. 34, 731–746 (2022).
Tyrakis, P. A. et al. S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 540, 236–241 (2016).
Piccirillo, A. R., Cattley, R. T., D’Cruz, L. M. & Hawse, W. F. Histone acetyltransferase CBP is critical for conventional effector and memory T-cell differentiation in mice. J. Biol. Chem. 294, 2397–2406 (2019).
Kakaradov, B. et al. Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat. Immunol. 18, 422–432 (2017).
Gray, S. M., Amezquita, R. A., Guan, T., Kleinstein, S. H. & Kaech, S. M. Polycomb repressive complex 2-mediated chromatin repression guides effector CD8+ T cell terminal differentiation and loss of multipotency. Immunity 46, 596–608 (2017).
Pace, L. et al. The epigenetic control of stemness in CD8+ T cell fate commitment. Science 359, 177–186 (2018).
Lau, C. M. et al. Epigenetic control of innate and adaptive immune memory. Nat. Immunol. 19, 963–972 (2018).
Sciume, G. et al. Rapid enhancer remodeling and transcription factor repurposing enable high magnitude gene induction upon acute activation of NK cells. Immunity 53, 745–758 (2020).
Mujal, A. M., Delconte, R. B. & Sun, J. C. Natural killer cells: from innate to adaptive features. Annu. Rev. Immunol. 39, 417–447 (2021).
Larsen, S. B. et al. Establishment, maintenance, and recall of inflammatory memory. Cell Stem Cell 28, 1758–1774 (2021).
Vierbuchen, T. et al. AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Mol. Cell 68, 1067–1082 (2017).
Wolf, B. K. et al. Cooperation of chromatin remodeling SWI/SNF complex and pioneer factor AP-1 shapes 3D enhancer landscapes. Nat. Struct. Mol. Biol. 30, 10–21 (2023).
Chandra, A. et al. Quantitative control of Ets1 dosage by a multi-enhancer hub promotes TH1 cell differentiation and protects from allergic inflammation. Immunity 56, 1451–1467 (2023).
Pham, D. et al. Batf pioneers the reorganization of chromatin in developing effector T cells via Ets1-dependent recruitment of Ctcf. Cell Rep. 29, 1203–1220 (2019).
Santosa, E. K., Lau, C. M., Sahin, M., Leslie, C. S. & Sun, J. C. 3D Chromatin dynamics during innate and adaptive immune memory acquisition. Preprint at bioRxiv https://doi.org/10.1101/2023.01.16.524322 (2023).
Gordon, S. M. et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55–67 (2012).
Intlekofer, A. M. et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6, 1236–1244 (2005).
Joshi, N. S. et al. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007).
Klose, C. S. N. et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157, 340–356 (2014).
Roychoudhuri, R. et al. BACH2 regulates CD8+ T cell differentiation by controlling access of AP-1 factors to enhancers. Nat. Immunol. 17, 851–860 (2016).
Omilusik, K. D. et al. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med. 212, 2027–2039 (2015).
Xin, A. et al. A molecular threshold for effector CD8+ T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat. Immunol. 17, 422–432 (2016).
Yang, C. Y. et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12, 1221–1229 (2011).
Zhou, X. et al. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity 33, 229–240 (2010).
van Helden, M. J. et al. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J. Exp. Med. 212, 2015–2025 (2015).
Kallies, A. et al. A role for Blimp1 in the transcriptional network controlling natural killer cell maturation. Blood 117, 1869–1879 (2011).
Jeevan-Raj, B. et al. The transcription factor Tcf1 contributes to normal NK cell development and function by limiting the expression of granzymes. Cell Rep. 20, 613–626 (2017).
Delconte, R. B. et al. The helix-loop-helix protein ID2 governs NK cell fate by tuning their sensitivity to interleukin-15. Immunity 44, 103–115 (2016).
Imianowski, C. J. et al. BACH2 restricts NK cell maturation and function, limiting immunity to cancer metastasis. J. Exp. Med. 219, e20211476 (2022).
Li, S. et al. The transcription factor Bach2 negatively regulates murine natural killer cell maturation and function. eLife 11, e77294 (2022).
Holmes, T. D. et al. The transcription factor Bcl11b promotes both canonical and adaptive NK cell differentiation. Sci. Immunol. 6, eabc9801 (2021).
Kallal, L. E. & Biron, C. A. Changing partners at the dance: variations in STAT concentrations for shaping cytokine function and immune responses to viral infections. JAKSTAT 2, e23504 (2013).
Li, P. et al. BATF–JUN is critical for IRF4-mediated transcription in T cells. Nature 490, 543–546 (2012).
Man, K. et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol. 14, 1155–1165 (2013).
Schraml, B. U. et al. The AP-1 transcription factor Batf controls TH17 differentiation. Nature 460, 405–409 (2009).
Kurachi, M. et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol. 15, 373–383 (2014).
Yao, S. et al. Interferon regulatory factor 4 sustains CD8+ T cell expansion and effector differentiation. Immunity 39, 833–845 (2013).
Santosa, E. K. et al. Control of nutrient uptake by IRF4 orchestrates innate immune memory. Nat. Immunol. https://doi.org/10.1038/s41590-023-01620-z (2023).
Glasmacher, E. et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1–IRF complexes. Science 338, 975–980 (2012).
Adams, N. M. et al. Transcription factor IRF8 orchestrates the adaptive natural killer cell response. Immunity 48, 1172–1182 (2018).
Beaulieu, A. M., Zawislak, C. L., Nakayama, T. & Sun, J. C. The transcription factor Zbtb32 controls the proliferative burst of virus-specific natural killer cells responding to infection. Nat. Immunol. 15, 546–553 (2014).
Rapp, M. et al. Core-binding factor beta and Runx transcription factors promote adaptive natural killer cell responses. Sci. Immunol. 2, eaan3796 (2017).
Cruz-Guilloty, F. et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 206, 51–59 (2009).
Wang, D. et al. The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity 48, 659–674 (2018).
Milner, J. J. et al. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257 (2017).
Sun, J. C. & Lanier, L. L. NK cell development, homeostasis and function: parallels with CD8+ T cells. Nat. Rev. Immunol. 11, 645–657 (2011).
Masopust, D. & Soerens, A. G. Tissue-resident T cells and other resident leukocytes. Annu Rev. Immunol. 37, 521–546 (2019).
Vivier, E. et al. Innate lymphoid cells: 10 years on. Cell 174, 1054–1066 (2018).
Huang, Y. et al. IL-25-responsive, lineage-negative KLRG1hi cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat. Immunol. 16, 161–169 (2015).
Huang, Y. et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 359, 114–119 (2018).
Mackay, L. K. et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016).
Mackay, L. K. et al. T-box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity 43, 1101–1111 (2015).
Schuster, I. S. et al. Infection induces tissue-resident memory NK cells that safeguard tissue health. Immunity 56, 531–546 e536 (2023).
Krmpotic, A., Bubic, I., Polic, B., Lucin, P. & Jonjic, S. Pathogenesis of murine cytomegalovirus infection. Microbes Infect. 5, 1263–1277 (2003).
Crough, T. & Khanna, R. Immunobiology of human cytomegalovirus: from bench to bedside. Clin. Microbiol Rev. 22, 76–98 (2009).
Kirchhammer, N. et al. NK cells with tissue-resident traits shape response to immunotherapy by inducing adaptive antitumor immunity. Sci. Transl. Med. 14, eabm9043 (2022).
Brownlie, D. et al. Expansions of adaptive-like NK cells with a tissue-resident phenotype in human lung and blood. Proc. Natl Acad. Sci. USA 118, e2016580118 (2021).
Berrien-Elliott, M. M. et al. Hematopoietic cell transplantation donor-derived memory-like NK cells functionally persist after transfer into patients with leukemia. Sci. Transl. Med. 14, eabm1375 (2022).
Marin, N. D. et al. Memory-like differentiation enhances NK cell responses to melanoma. Clin. Cancer Res. 27, 4859–4869 (2021).
Romee, R. et al. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci. Transl. Med. 8, 357ra123 (2016).
Cichocki, F. et al. Adaptive NK cell reconstitution is associated with better clinical outcomes. JCI Insight 4, e125553 (2019).
Boudreau, J. E. et al. KIR3DL1/HLA-B subtypes govern acute myelogenous leukemia relapse after hematopoietic cell transplantation. J. Clin. Oncol. 35, 2268–2278 (2017).
Liu, E. et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 382, 545–553 (2020).
Gauthier, L. et al. Control of acute myeloid leukemia by a trifunctional NKp46-CD16a-NK cell engager targeting CD123. Nat. Biotechnol. https://doi.org/10.1038/s41587-022-01626-2 (2023).
Jacquelot, N., Seillet, C., Vivier, E. & Belz, G. T. Innate lymphoid cells and cancer. Nat. Immunol. 23, 371–379 (2022).
Acknowledgements
The authors apologize to all those whose significant and valuable contributions could not be cited or discussed in this manuscript owing to space limitations. We thank members of the Sun lab for helpful feedback on the manuscript. J.C.S. was supported by the Ludwig Center for Cancer Immunotherapy, the American Cancer Society, the Burroughs Wellcome Fund, and the NIH (AI100874, AI130043, AI155558, and P30CA008748).
Author information
Authors and Affiliations
Contributions
J.C.S. and E.K.S. contributed equally to all aspects of this article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Jeffrey Miller and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editor: Jamie D. K. Wilson, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Santosa, E.K., Sun, J.C. Cardinal features of immune memory in innate lymphocytes. Nat Immunol 24, 1803–1812 (2023). https://doi.org/10.1038/s41590-023-01607-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-023-01607-w