Abstract
Naive CD4+ T lymphocytes initially undergo antigen-specific activation to promote a broad-spectrum response before adopting bespoke cytokine expression profiles shaped by intercellular microenvironmental cues, resulting in pathogen-focused modular cytokine responses. Interleukin (IL)-4-induced Gata3 upregulation is important for the helper type 2 T cell (TH2 cell) polarization associated with anti-helminth immunity and misdirected allergic inflammation. Whether additional microenvironmental factors participate is unclear. Using whole mouse-genome CRISPR–Cas9 screens, we discovered a previously unappreciated role for αvβ3 integrin in TH2 cell differentiation. Low-level αvβ3 expression by naive CD4+ T cells contributed to pan-T cell activation by promoting T–T cell clustering and IL-2/CD25/STAT5 signaling. Subsequently, IL-4/Gata3-induced selective upregulation of αvβ3 licensed intercellular αvβ3–Thy1 interactions among TH2 cells, enhanced mammalian target of rapamycin (mTOR) signaling, supported differentiation and promoted IL-5/IL-13 production. In mice, αvβ3 was required for efficient, allergen-driven, antigen-specific lung TH2 cell responses. Thus, αvβ3-expressing TH2 cells form multicellular factories to propagate and amplify TH2 cell responses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All high-throughput data in the present study were deposited at the Gene Expression Omnibus under accession no. GSE179210. TH cell transcriptomic data were obtained from Th-express (https://th-express.org). Source data are provided with this paper.
References
Paul, W. E. & Zhu, J. How are TH2-type immune responses initiated and amplified? Nat. Rev. Immunol. 10, 225–235 (2010).
Walker, J. A. & McKenzie, A. N. J. TH2 cell development and function. Nat. Rev. Immunol. 18, 121–133 (2018).
van Panhuys, N., Klauschen, F. & Germain, R. N. T-cell-receptor-dependent signal intensity dominantly controls CD4+ T cell polarization in vivo. Immunity 41, 63–74 (2014).
Zhu, J. Transcriptional regulation of Th2 cell differentiation. Immunol. Cell Biol. 88, 244–249 (2010).
Luksch, C. R. et al. Intercellular adhesion molecule-1 inhibits interleukin 4 production by naive T cells. Proc. Natl Acad. Sci. USA 96, 3023–3028 (1999).
Salomon, B. & Bluestone, J. A. LFA-1 interaction with ICAM-1 and ICAM-2 regulates Th2 cytokine production. J. Immunol. 161, 5138–5142 (1998).
Smits, H. H. et al. Intercellular adhesion molecule-1/LFA-1 ligation favors human Th1 development. J. Immunol. 168, 1710–1716 (2002).
Sabatos, C. A. et al. A synaptic basis for paracrine interleukin-2 signaling during homotypic T cell interaction. Immunity 29, 238–248 (2008).
Henriksson, J. et al. Genome-wide CRISPR screens in T helper cells reveal pervasive crosstalk between activation and differentiation. Cell 176, 882–896.e818 (2019).
Peng, H. et al. Monocyte chemotactic protein-induced protein 1 controls allergic airway inflammation by suppressing IL-5-producing TH2 cells through the Notch/Gata3 pathway. J. Allergy Clin. Immunol. 142, 582–594.e510 (2018).
Zhu, J. & Paul, W. E. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol. Rev. 238, 247–262 (2010).
Tur-Gracia, S. & Martinez-Quiles, N. Emerging functions of cytoskeletal proteins in immune diseases. J. Cell Sci. 134, jcs253534 (2021).
Sun, Z., Costell, M. & Fässler, R. Integrin activation by talin, kindlin and mechanical forces. Nat. Cell Biol. 21, 25–31 (2019).
Pribila, J. T., Quale, A. C., Mueller, K. L. & Shimizu, Y. Integrins and T cell-mediated immunity. Annu. Rev. Immunol. 22, 157–180 (2004).
Sorcini, D. et al. Wnt/beta-catenin signaling induces integrin alpha4beta1 in T cells and promotes a progressive neuroinflammatory disease in mice. J. Immunol. 199, 3031–3041 (2017).
Halim, T. Y. et al. Group 2 innate lymphoid cells license dendritic cells to potentiate memory TH2 cell responses. Nat. Immunol. 17, 57–64 (2016).
Verma, N. K. et al. LFA-1/ICAM-1 ligation in human T cells promotes Th1 polarization through a GSK3beta signaling-dependent notch pathway. J. Immunol. 197, 108–118 (2016).
Ross, S. H. & Cantrell, D. A. Signaling and function of interleukin-2 in T lymphocytes. Annu. Rev. Immunol. 36, 411–433 (2018).
Zheng, W. & Flavell, R. A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 (1997).
Verschelde, C. et al. A1/Bfl-1 expression is restricted to TCR engagement in T lymphocytes. Cell Death Differ. 10, 1059–1067 (2003).
Kurebayashi, Y. et al. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORgamma. Cell Rep. 1, 360–373 (2012).
Lin, W. et al. RUNX1/EGFR pathway contributes to STAT3 activation and tumor growth caused by hyperactivated mTORC1. Mol. Ther. Oncolytics 23, 387–401 (2021).
Nowyhed, H. N. et al. The nuclear receptor nr4a1 controls CD8 T cell development through transcriptional suppression of runx3. Sci. Rep. 5, 9059 (2015).
Chapman, N. M. & Houtman, J. C. Functions of the FAK family kinases in T cells: beyond actin cytoskeletal rearrangement. Immunol. Res. 59, 23–34 (2014).
Rashmi et al. ZINC40099027 activates human focal adhesion kinase by accelerating the enzymatic activity of the FAK kinase domain. Pharm. Res. Perspect. 9, e00737 (2021).
Wang, Q., More, S. K., Vomhof-DeKrey, E. E., Golovko, M. Y. & Basson, M. D. Small molecule FAK activator promotes human intestinal epithelial monolayer wound closure and mouse ulcer healing. Sci. Rep. 9, 14669 (2019).
Humphries, J. D., Byron, A. & Humphries, M. J. Integrin ligands at a glance. J. Cell Sci. 119, 3901–3903 (2006).
Beissert, S. et al. Impaired cutaneous immune responses in Thy-1-deficient mice. J. Immunol. 161, 5296–5302 (1998).
Morris, R. J. Thy-1, a pathfinder protein for the post-genomic era. Front. Cell Dev. Biol. 6, 173 (2018).
Parnas, O. et al. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162, 675–686 (2015).
Peng, M. et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016).
Wellen, K. E. et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009).
Kerscher, B. et al. BET bromodomain inhibitor iBET151 impedes human ILC2 activation and prevents experimental allergic lung inflammation. Front. Immunol. 10, 678 (2019).
Bandukwala, H. S. et al. Selective inhibition of CD4+ T-cell cytokine production and autoimmunity by BET protein and c-Myc inhibitors. Proc. Natl Acad. Sci. USA 109, 14532–14537 (2012).
Lehnertz, B. et al. Activating and inhibitory functions for the histone lysine methyltransferase G9a in T helper cell differentiation and function. J. Exp. Med. 207, 915–922 (2010).
Djuretic, I. M. et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8, 145–153 (2007).
Kohu, K. et al. The Runx3 transcription factor augments Th1 and down-modulates Th2 phenotypes by interacting with and attenuating GATA3. J. Immunol. 183, 7817–7824 (2009).
Komine, O. et al. The Runx1 transcription factor inhibits the differentiation of naive CD4+ T cells into the Th2 lineage by repressing GATA3 expression. J. Exp. Med. 198, 51–61 (2003).
Naoe, Y. et al. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J. Exp. Med. 204, 1749–1755 (2007).
Malik, N. et al. The transcription factor CBFB suppresses breast cancer through orchestrating translation and transcription. Nat. Commun. 10, 2071 (2019).
Overstreet, M. G. et al. Inflammation-induced interstitial migration of effector CD4+ T cells is dependent on integrin alphaV. Nat. Immunol. 14, 949–958 (2013).
Schrock, D. C. et al. Pivotal role for alphaV integrins in sustained Tfh support of the germinal center response for long-lived plasma cell generation. Proc. Natl Acad. Sci. USA 116, 4462–4470 (2019).
Gaylo-Moynihan, A. et al. Programming of distinct chemokine-dependent and -independent search strategies for Th1 and Th2 cells optimizes function at inflamed sites. Immunity 51, 298–309.e296 (2019).
Zenke, S. et al. Quorum regulation via nested antagonistic feedback circuits mediated by the receptors CD28 and CTLA-4 confers robustness to T cell population dynamics. Immunity 52, 313–327.e317 (2020).
Wu, C., You, J., Fu, J., Wang, X. & Zhang, Y. Phosphatidylinositol 3-kinase/Akt mediates integrin signaling to control RNA polymerase I transcriptional activity. Mol. Cell Biol. 36, 1555–1568 (2016).
Paul, R. et al. FAK activates AKT-mTOR signaling to promote the growth and progression of MMTV-Wnt1-driven basal-like mammary tumors. Breast Cancer Res. 22, 59 (2020).
Lohoff, M. et al. Early growth response protein-1 (Egr-1) is preferentially expressed in T helper type 2 (Th2) cells and is involved in acute transcription of the Th2 cytokine interleukin-4. J. Biol. Chem. 285, 1643–1652 (2010).
Li, B. et al. The early growth response factor-1 contributes to interleukin-13 production by mast cells in response to stem cell factor stimulation. J. Immunotoxicol. 5, 163–171 (2008).
Minutti, C. M. et al. Epidermal growth factor receptor expression licenses type-2 helper T cells to function in a T cell receptor-independent fashion. Immunity 47, 710–722 e716 (2017).
Roy, S. et al. EGFR-HIF1alpha signaling positively regulates the differentiation of IL-9 producing T helper cells. Nat. Commun. 12, 3182 (2021).
Smit, L. A. et al. Mold allergen sensitization in adult asthma according to integrin β3 polymorphisms and Toll-like receptor 2/+596 genotype. J. Allergy Clin. Immunol. 128, 185–191.e187 (2011).
Weiss, L. A. et al. Variation in ITGB3 is associated with asthma and sensitization to mold allergen in four populations. Am. J. Respir. Crit. Care Med. 172, 67–73 (2005).
Han, Y. et al. Genome-wide analysis highlights contribution of immune system pathways to the genetic architecture of asthma. Nat. Commun. 11, 1776 (2020).
Izuhara, K. et al. Periostin in allergic inflammation. Allergol. Int. 63, 143–151 (2014).
Platt, R. J. et al. CRISPR–Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Barlow, J. L. et al. Innate IL-13-producing nuocytes arise during allergic lung inflammation and contribute to airways hyperreactivity. J. Allergy Clin. Immunol. 129, 191–198.e191-194 (2012).
Lacy-Hulbert, A. et al. Ulcerative colitis and autoimmunity induced by loss of myeloid alphav integrins. Proc. Natl Acad. Sci. USA 104, 15823–15828 (2007).
Morgan, E. A. et al. Dissection of platelet and myeloid cell defects by conditional targeting of the beta3-integrin subunit. FASEB J. 24, 1117–1127 (2010).
Aghajani, K., Keerthivasan, S., Yu, Y. & Gounari, F. Generation of CD4CreER(T2) transgenic mice to study development of peripheral CD4-T-cells. Genesis 50, 908–913 (2012).
Chu, V. T. et al. Efficient CRISPR-mediated mutagenesis in primary immune cells using CrispRGold and a C57BL/6 Cas9 transgenic mouse line. Proc. Natl Acad. Sci. USA 113, 12514–12519 (2016).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Schmidl, C., Rendeiro, A. F., Sheffield, N. C. & Bock, C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 12, 963–965 (2015).
Acknowledgements
We thank the ARES staff, genotyping facility, flow cytometry core and the National Institute for Health and Care Research, Cambridge Biomedical Research Centre Cell Phenotyping hub for their technical assistance. Funding: the present study was supported by grants from the UK Medical Research Council (MRC; grant no. U105178805) and Wellcome Trust (grant nos. 100963/Z/13/Z and 220223/Z/20/Z). A.C.H.S. was supported by the Croucher Foundation. M.D.K. was supported by an MRC CARP award (MR/T005386/1). We thank the National Institutes of Health Tetramer Core Facility for supplying the 2W1S-MHC-II tetramer and D. Withers for providing the Cd4CreERT2 mice.
Author information
Authors and Affiliations
Contributions
A.C.H.S. designed and performed experiments and wrote the paper. A.C.F.F., J.M., P.A.C., M.W.D.H., A.C., M.S., H.E.J., P.K. and M.D.K. performed experiments, provided advice on experimental design and interpretation, and commented on the manuscript. A.N.J.M. supervised the project, designed the experiments and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: S. Houston in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Optimisation of a genome-wide screen for regulators of TH2 cell differentiation.
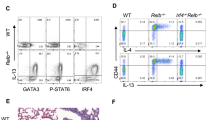
Extended Data Fig. 1 (a) Schematic of the optimised TH2 cell culture protocol for CRISPR screening. (b) Flow cytometric analysis of IL-13Tom expression by TH2 cells transduced with NT or Gata3 sgRNAs using the optimised protocol. Data representative of 3 independent experiments. (c) RNA-sequencing analysis of Gata3-targeted versus non-targeted TH2 cells using the optimised screening protocol. (d) KEGG pathway analysis of genes downregulated in Gata3-targeted versus non-targeted TH2 cells. (e) and (f) Gene set enrichment analyses of genome-wide positive regulators of TH2 cell differentiation identified in the screen. (g) Selection of the top 1018 genes from the genome-wide screen for a secondary screen (fold change > 0.06 and p-value < 0.07). (h) Validation of novel regulators by individual confirmatory sgRNA knockdown. Data representative of 3 independent experiments; mean ± SD; one-way ANOVA with Dunnett’s post-hoc test. (i) Validation of novel regulators as in (h) with corresponding TH1 comparisons. (h) & (i) **** P < 0.0001, ***P = 0.0006 (Fermt3), **P = 0.0018 (Hsp90b1), 0.0053 (Tfap4), *P = 0.0102 (Fnta), 0.0169 (Smarcc1), 0.0212 (Apbb1ip), 0.0252 (Kmt2d).
Extended Data Fig. 2 Differential integrin expression by in vitro and in vivo TH cells.
Extended Data Fig. 2(a) Flow cytometric analysis of αv and β3 expression by TH2 cells in vitro. (b) Flow cytometric analysis of α4 (Itga4) and β1 (Itgb1) expression by TH cells in vitro. Data are representative of 2 independent experiments with 4 biologically independent samples in each experiment; unpaired two-sided t-test; ****P < 0.0001, ***P = 0.0004. (c) Gating strategy for TH cell subsets in the papain-challenged lung. (d) Flow cytometric gating strategy and quantification of lymphoid populations in naive mice. (e) Flow cytometric analysis of αv and β3 expression in control, αv- or β3- deficient naïve CD4+ T cells. (f) and (g) Quantification of lymphoid populations in naive mice. Data representative of 2 independent experiments with 3 biologically independent samples in each experiment; mean ± SD; unpaired two-sided t-test. (h) Schematic of the experimental induction of type 2 inflammation in the mouse lung with OVA/Alum. (i) Schematic of the experimental induction of type 2 inflammation in the mouse lung with papain. (j) Schematic of the experimental induction of type 1 inflammation in the mouse lung with LPS.
Extended Data Fig. 3 Gating strategy of TH cell populations in vivo.
Extended Data Fig. 3(a) & (b) Flow cytometric gating strategy for cytokine- and transcription factor- expressing THeff cells in the mediastinal lymph node of OVA/Alum-challenged mice. (c) Flow cytometric gating strategy for 2W1S-tetramer-specific TH2 cells in the papain-challenged lung. (d) & (e) Flow cytometric gating and quantification of 2W1S-tetramer-specific IL-5 and IL-13 producing TH effector cells in PMA/ionomycin stimulated lung lymphocytes. Data are pooled from 2 independent experiments and represent mean ± SD (n = 6 mice in naïve and papain only control groups, n = 16 mice in Cd4Cre group, n = 15 mice in ItgavCD4KO group); unpaired two-sided t-test; *P = 0.0418 (top, IL-5) and 0.0211 (bottom, IL-13). (f) Flow cytometric gating strategy for IFN-γ expressing THeff cells in the mediastinal lymph node of LPS-challenged mice.
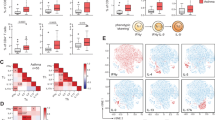
Extended Data Fig. 4 Genome-wide transcriptomic analysis of av-deficient TH cells.
Extended Data Fig. 4(a) Principal component analysis of the transcriptomes of control versus αv-deficient TH cells. (b) Volcano plot depiction of differentially expressed genes in control versus αv-deficient TH2 cells. (c) & (d) RNA expression of type 2 genes by control or αv-deficient TH cells; Il13 ***P = 0.0002, **P = 0.0012; Il5 ***P = 0.0006, **P = 0.0025; Il4 **P = 0.0046, *P = 0.0167, Gata3 ****P < 0.0001. (e) RNA expression of type-1 signalling genes by control or αv-deficient TH cells; Stat1 ****P < 0.0001, *P = 0.0438; Isg20 *P = 0.0148; Runx3 **P = 0.0052, *P = 0.0220. (c) - (e) mean ± SEM; one-way ANOVA with Tukey’s post-hoc test. (f) Flow cytometric analysis of Runx3 expression by αv-deficient TH cells. Data are representative of 2 independent experiments; unpaired two-sided t-test; ***P = 0.0003. (g) KEGG pathway analysis of genes downregulated in αv-deficient versus control TH2 cells.
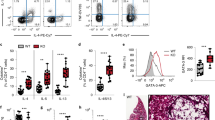
Extended Data Fig. 5 avb3-Thy1 inhibition does not affect TH1 cell differentiation and IFN-g expression.
Extended Data Fig. 5(a) Flow cytometric analysis of cytokine expression by TH1 cells cultured in the presence of vehicle (DMSO) or cilengitide. Data are representative of 3 independent experiments with 5 biologically independent samples in each experiment; mean ± SD; unpaired two-sided t-test; ***P = 0.0001 (top, % IFN-γ + cells) and 0.0005 (bottom, IFN-γ MFI). (b) Flow cytometric analysis of cytokine expression by TH1 cells cultured in the presence of isotype or anti-αv antibody. Data are representative of 2 independent experiments with 5 biologically independent samples in each experiment; mean ± SD; unpaired two-sided t-test; ****P < 0.0001. (c) Flow cytometric analysis of cytokine expression by TH1 cells cultured in the presence of isotype or anti-β3 antibody. Data are representative of 2 independent experiments with 5 biologically independent samples in each experiment; mean ± SD; unpaired two-sided t-test. (d) Flow cytometric analysis of cytokine expression by TH1 cells cultured in the presence of isotype or anti-Thy1 antibody. Data are representative of 2 independent experiments; paired two-sided t-test. (e) Flow cytometric analysis of Thy1-Fc conjugation to epoxy beads. (f) Flow cytometric gating strategy of CD4 T-T doublets in the mediastinal lymph nodes of OVA/Alum challenged mice. (g) Flow cytometric quantification of CD4 T-T doublets as in (f); *P = 0.0132 (Cd4Cre vs ItgavCD4KO), 0.0248 (Cd4Cre vs Itgb3CD4KO). (h) Flow cytometric quantification of type-2 cytokine expressing-CD4 T-T doublets as in (f); IL-5 + CD4 doublet: **P = 0.0021, *P = 0.0129; IL-13 + CD4 doublet: **P = 0.0046, *P = 0.0148. (g) – (h) data are pooled from 2 independent experiments and represent mean ± SD (n = 12 mice in Cd4Cre and ItgavCD4KO groups, n = 11 mice in Itgb3CD4KO group); one-way ANOVA with Dunnett’s post-hoc test.
Extended Data Fig. 6 Proposed model for αvβ3-mediated potentiation of TH2 cell responses.
Naïve CD4+ T cells express low levels of integrin αvβ3 which contribute to T cell activation and signalling via the IL-2/CD25/STAT5 axis. IL-4-mediated Gata3 induction upregulates αvβ3 during TH2 cell differentiation, permitting intercellular interactions among TH2 cells via αvβ3-Thy1 binding. Such interactions enhance mTOR signalling and support optimal TH2 responses in vivo.
Supplementary information
Supplementary Table 1
Analyses of genome-wide and secondary screens by Wilcoxon-based hit calling.
Source data
Source Data Fig. 6
Unprocessed immunmoblot of Thy1 detection.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Szeto, A.C.H., Ferreira, A.C.F., Mannion, J. et al. An αvβ3 integrin checkpoint is critical for efficient TH2 cell cytokine polarization and potentiation of antigen-specific immunity. Nat Immunol 24, 123–135 (2023). https://doi.org/10.1038/s41590-022-01378-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-022-01378-w
This article is cited by
-
CRISPR–Cas9 applications in T cells and adoptive T cell therapies
Cellular & Molecular Biology Letters (2024)