Abstract
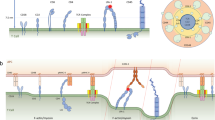
The T cell antigen receptor (TCR) expressed on thymocytes interacts with self-peptide major histocompatibility complex (pMHC) ligands to signal apoptosis or survival. Here, we found that negative-selection ligands induced thymocytes to exert forces on the TCR and the co-receptor CD8 and formed cooperative TCR–pMHC–CD8 trimolecular ‘catch bonds’, whereas positive-selection ligands induced less sustained thymocyte forces on TCR and CD8 and formed shorter-lived, independent TCR–pMHC and pMHC–CD8 bimolecular ‘slip bonds’. Catch bonds were not intrinsic to either the TCR–pMHC or the pMHC–CD8 arm of the trans (cross-junctional) heterodimer but resulted from coupling of the extracellular pMHC–CD8 interaction to the intracellular interaction of CD8 with TCR–CD3 via associated kinases to form a cis (lateral) heterodimer capable of inside-out signaling. We suggest that the coupled trans–cis heterodimeric interactions form a mechanotransduction loop that reinforces negative-selection signaling that is distinct from positive-selection signaling in the thymus.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Rudolph, M. G., Stanfield, R. L. & Wilson, I. A. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466 (2006).
Stepanek, O. et al. Coreceptor scanning by the T cell receptor provides a mechanism for T cell tolerance. Cell 159, 333–345 (2014).
Palmer, E. & Naeher, D. Affinity threshold for thymic selection through a T-cell receptor-co-receptor zipper. Nat. Rev. Immunol. 9, 207–213 (2009).
Daniels, M. A. et al. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444, 724–729 (2006).
Huang, J. et al. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature 464, 932–936 (2010).
Huppa, J. B. et al. TCR–peptide–MHC interactions in situ show accelerated kinetics and increased affinity. Nature 463, 963–967 (2010).
Jiang, N. et al. Two-stage cooperative T cell receptor-peptide major histocompatibility complex-CD8 trimolecular interactions amplify antigen discrimination. Immunity 34, 13–23 (2011).
Martinez, R. J., Andargachew, R., Martinez, H. A. & Evavold, B. D. Low-affinity CD4+ T cells are major responders in the primary immune response. Nat. Commun. 7, 13848 (2016).
Liu, B., Chen, W., Evavold, B. D. & Zhu, C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 157, 357–368 (2014).
Hong, J. et al. Force-regulated in situ TCR-peptide-bound MHC class II kinetics determine functions of CD4+ T cells. J. Immunol. 195, 3557–3564 (2015).
Das, D. K. et al. Force-dependent transition in the T-cell receptor β-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc. Natl. Acad. Sci. USA 112, 1517–1522 (2015).
Liu, Y. et al. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc. Natl. Acad. Sci. USA 113, 5610–5615 (2016).
Sarangapani, K. K., Marshall, B. T., McEver, R. P. & Zhu, C. Molecular stiffness of selectins. J. Biol. Chem. 286, 9567–9576 (2011).
Erman, B. et al. Coreceptor signal strength regulates positive selection but does not determine CD4/CD8 lineage choice in a physiologic in vivo model. J. Immunol. 177, 6613–6625 (2006).
Hui, E. & Vale, R. D. In vitro membrane reconstitution of the T-cell receptor proximal signaling network. Nat. Struct. Mol. Biol. 21, 133–142 (2014).
Fiore, V. F., Ju, L., Chen, Y., Zhu, C. & Barker, T. H. Dynamic catch of a Thy-1–α5β1+syndecan-4 trimolecular complex. Nat. Commun. 5, 4886 (2014).
Chen, W., Lou, J., Evans, E. A. & Zhu, C. Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. J. Cell Biol. 199, 497–512 (2012).
Bashour, K. T. et al. CD28 and CD3 have complementary roles in T-cell traction forces. Proc. Natl. Acad. Sci. USA 111, 2241–2246 (2014).
Zhang, Y., Ge, C., Zhu, C. & Salaita, K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat. Commun. 5, 5167 (2014).
Hogquist, K. A. et al. Identification of a naturally occurring ligand for thymic positive selection. Immunity 6, 389–399 (1997).
Santori, F. R. et al. Rare, structurally homologous self-peptides promote thymocyte positive selection. Immunity 17, 131–142 (2002).
Ju, L., Dong, J. F., Cruz, M. A. & Zhu, C. The N-terminal flanking region of the A1 domain regulates the force-dependent binding of von Willebrand factor to platelet glycoprotein Ibα. J. Biol. Chem. 288, 32289–32301 (2013).
Dustin, M. L. T-cell activation through immunological synapses and kinapses. Immunol. Rev. 221, 77–89 (2008).
Chen, W., Evans, E. A., McEver, R. P. & Zhu, C. Monitoring receptor-ligand interactions between surfaces by thermal fluctuations. Biophys. J. 94, 694–701 (2008).
Chesla, S. E., Selvaraj, P. & Zhu, C. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys. J. 75, 1553–1572 (1998).
Dushek, O. & van der Merwe, P. A. An induced rebinding model of antigen discrimination. Trends Immunol. 35, 153–158 (2014).
Hwang, S. et al. TCR ITAM multiplicity is required for the generation of follicular helper T-cells. Nat. Commun. 6, 6982 (2015).
Hwang, S. et al. Reduced TCR signaling potential impairs negative selection but does not result in autoimmune disease. J. Exp. Med. 209, 1781–1795 (2012).
Nika, K. et al. Constitutively active Lck kinase in T cells drives antigen receptor signal transduction. Immunity 32, 766–777 (2010).
Thome, M., Duplay, P., Guttinger, M. & Acuto, O. Syk and ZAP-70 mediate recruitment of p56lck/CD4 to the activated T cell receptor/CD3/zeta complex. J. Exp. Med. 181, 1997–2006 (1995).
Manz, B. N. et al. Small molecule inhibition of Csk alters affinity recognition by T cells. eLife 4, e08088 (2015).
Kinashi, T. Intracellular signalling controlling integrin activation in lymphocytes. Nat. Rev. Immunol. 5, 546–559 (2005).
Yachi, P. P., Ampudia, J., Zal, T. & Gascoigne, N. R. Altered peptide ligands induce delayed CD8-T cell receptor interaction: a role for CD8 in distinguishing antigen quality. Immunity 25, 203–211 (2006).
Arcaro, A. et al. CD8beta endows CD8 with efficient coreceptor function by coupling T cell receptor/CD3 to raft-associated CD8/p56(lck) complexes. J. Exp. Med. 194, 1485–1495 (2001).
Doucey, M. A. et al. CD3 delta establishes a functional link between the T cell receptor and CD8. J. Biol. Chem. 278, 3257–3264 (2003).
Thome, M., Germain, V., DiSanto, J. P. & Acuto, O. The p56lck SH2 domain mediates recruitment of CD8/p56lck to the activated T cell receptor/CD3/zeta complex. Eur. J. Immunol. 26, 2093–2100 (1996).
Straus, D. B., Chan, A. C., Patai, B. & Weiss, A. SH2 domain function is essential for the role of the Lck tyrosine kinase in T cell receptor signal transduction. J. Biol. Chem. 271, 9976–9981 (1996).
Lo, W. L. et al. Lck promotes Zap70-dependent LAT phosphorylation by bridging Zap70 to LAT. Nat. Immunol. 19, 733–741 (2018).
Goldrath, A. W., Hogquist, K. A. & Bevan, M. J. CD8 lineage commitment in the absence of CD8. Immunity 6, 633–642 (1997).
Lou, J. & Zhu, C. A structure-based sliding-rebinding mechanism for catch bonds. Biophys. J. 92, 1471–1485 (2007).
Alam, S. M. et al. T-cell-receptor affinity and thymocyte positive selection. Nature 381, 616–620 (1996).
Artyomov, M. N., Lis, M., Devadas, S., Davis, M. M. & Chakraborty, A. K. CD4 and CD8 binding to MHC molecules primarily acts to enhance Lck delivery. Proc. Natl. Acad. Sci. USA 107, 16916–16921 (2010).
Feng, Y. et al. Mechanosensing drives acuity of αβ T-cell recognition. Proc. Natl. Acad. Sci. USA 114, E8204–E8213 (2017).
Van Laethem, F., Tikhonova, A. N. & Singer, A. MHC restriction is imposed on a diverse T cell receptor repertoire by CD4 and CD8 co-receptors during thymic selection. Trends Immunol. 33, 437–441 (2012).
Le Borgne, M. et al. The impact of negative selection on thymocyte migration in the medulla. Nat. Immunol. 10, 823–830 (2009).
Sauer, K., Huang, Y. H., Lin, H., Sandberg, M. & Mayr, G. W. Phosphoinositide and inositol phosphate analysis in lymphocyte activation. Curr. Protoc. Immunol. 87, 11.11.1 (2009).
Puls, K. L., Hogquist, K. A., Reilly, N. & Wright, M. D. CD53, a thymocyte selection marker whose induction requires a lower affinity TCR-MHC interaction than CD69, but is up-regulated with slower kinetics. Int. Immunol. 14, 249–258 (2002).
Nathenson, S. G., Geliebter, J., Pfaffenbach, G. M. & Zeff, R. A. Murine major histocompatibility complex class-I mutants: molecular analysis and structure-function implications. Annu. Rev. Immunol. 4, 471–502 (1986).
Degano, M. et al. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity 12, 251–261 (2000).
Tallquist, M. D., Yun, T. J. & Pease, L. R. A single T cell receptor recognizes structurally distinct MHC/peptide complexes with high specificity. J. Exp. Med. 184, 1017–1026 (1996).
Huang, J., Edwards, L. J., Evavold, B. D. & Zhu, C. Kinetics of MHC-CD8 interaction at the T cell membrane. J. Immunol. 179, 7653–7662 (2007).
Marshall, B. T. et al. Direct observation of catch bonds involving cell-adhesion molecules. Nature 423, 190–193 (2003).
Hailman, E. & Allen, P. M. Inefficient cell spreading and cytoskeletal polarization by CD4+CD8+ thymocytes: regulation by the thymic environment. J. Immunol. 175, 4847–4857 (2005).
Hailman, E., Burack, W. R., Shaw, A. S., Dustin, M. L. & Allen, P. M. Immature CD4+CD8+ thymocytes form a multifocal immunological synapse with sustained tyrosine phosphorylation. Immunity 16, 839–848 (2002).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2017).
Acknowledgements
We thank L. Lawrence for maintaining the mouse colonies; L. Doudy for thymocyte purification; W. Chen and Z. Li for assistance with experiments and discussions; and the NIH Tetramer Core Facility at Emory University for providing the pMHC monomers. This work was supported by NIH grants CA214354 and AI124680 (to C.Z.), and NS071518 and AI096879 (to B.D.E.).
Author information
Authors and Affiliations
Contributions
J.H., C.G., K.S., B.D.E., A.S., and C.Z. designed experiments; J.H., C.G., P.J., Z.Y., B.L., K.B., and K.L. performed experiments; Y.Z. and K.S. provided DNA force probes; M.S. and A.S. provided the OT1.CD8.4 mice; A.P. and P.L. provided the OT1.6F mice; W.R. performed modeling analysis; C.G., Z.Y., and X.Y. performed statistical analyses; and J.H. and C.Z. analyzed the data and wrote the paper, to which other authors contributed.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Integrated supplementary information
Supplementary Figure 1 Lck inhibition eliminated trimolecular bonds of CD8.4 thymocytes with wQ4H7 but not with wQ4R7.
Related to Fig. 2. Molecular stiffness histograms of complexes of DMSO-treated OT1 (first column), Lck inhibitor-treated OT1.CD8.4 (second column) and DMSO-treated OT1.CD8.4 (third column) thymocytes with wQ4H7 (upper row) or wQ4R7 (bottom row). Data (bar) were fitted by a single or double Gaussian per panel. The fitting parameters and statistics for their comparisons are summarized in Supplementary Tables 4a, b and 5a, respectively.
Supplementary Figure 2 Further confirmation of the thymocyte negative-selection criteria by force-dependent bond lifetimes, molecular stiffness, and cell pulling.
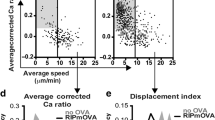
Related to Fig. 4. a, Mean ± s.e.m. of lifetime versus force plots of total bonds with indicated peptides presented by H2-Kb (green square and blue diamond) or H2-Kbm3 (red square), TCR bonds with three peptides presented by H2-Kbα3A2 (brown circle), and CD8 bond with VSV:H-2Kb (black triangle) measured using 2C TCR transgenic mice. The total bonds with the negative selecting ligands, SIYR:H-2Kb and dEV8:H-2Kbm3, exhibited catch bond behavior at low forces whereas those with the positive selecting ligands, dEV8, EVSV and p2Ca bound to H-2Kb, showed slip bond behavior9–11,52. Like mOVA, the super agonist SIYR also formed catch bond at low forces with the 2C TCR in the absence of CD8, but the other two pMHC bimolecular interactions and the MHC–CD8 interaction formed slip bonds. The numbers of bond lifetime measurements per curve for different ligands, the results of statistical tests examining the trends of the curves and their differences are summarized in Supplementary Tables 1d, 2c and 3a,b, respectively. b, Molecular stiffness histograms of 2C TCR bimolecular complexes with the indicated peptides bound to H-2Kbα3A2, total complexes with these peptides bound to H-2Kb, and total complexes with dEV8:H-2Kbm3 and p2Ca:H-2Kb. Data (bar) were fitted by a single (black curve) or double (black curve = cyan curve + red curve) Gaussian. The fitting parameters and statistics for their comparisons are summarized in Supplementary Tables 4c and 5c, respectively. c, Representative images of thymocytes placed on wVSV, wCatnb, and wOVA tagged with a 13.1 pN MTP viewed in the bright-field (left column), fluorescence (middle column), and merged (right column) channels. Scale bar = 5 µm. d, Comparison of initial force signals from wVSV, wCatnb, and wOVA tagged with a 13.1 pN MTP. e, Comparison of force signal decays of OT1 thymocytes pulled on mOVA and wOVA tagged with a 13.1 pN MTP. f, Representative images of thymocytes pulled on indicated peptides presented by H-2Kb tagged with a 4.7 pN MTP viewed in the bright-field (left column), fluorescence (middle column), and merged (right column) channels. Scale bar = 5 µm. g, Comparison of normalized fluorescence intensity (points, left ordinate) and fraction of positive cells (bars, right ordinate) for indicated peptides bound to H2-Kb. Normalized fluorescence intensity was calculated by dividing the mean fluorescence intensity from a Cy5 positive cell by the background. Data are presented as mean ± s.e.m of all positive cells (each cell is represented by a point with N indicating the number of positive cells). h, Comparison of force signal decays of OT1 thymocytes pulled on wQ4H7 and wQ4R7 tagged by a 4.7 pN MTP. i, Representative images of OT1 thymocytes pulled on wOVA tagged with a 13.1 pN MTP treated with the indicated agents: DMSO control, actin polymerization inhibitor latruculin A, and ROCK inhibitor Y-27632. OT1 thymocytes before (top row) and 10 min after (bottom row) the drug treatment were viewed in the bright-field (left column), fluorescence (middle column), and merged (right column) channels. Scale bar = 5 µm. j, Comparison of force signal decays of OT1 thymcoytes pulled on wOVA over 20 min for the same treatments as in (i). Data in (e,h,j) are presented as mean ± s.e.m. of fluorescence intensity normalized by the initial value at 0 min (N = number of cells pooled from ≥at least two independent experiments). *and **** denote p < 0.05 and 0.0001, respectively, by two-way ANOVA.
Supplementary Figure 3 2D binding at zero force does not provide clear criteria for thymocyte negative selection.
Related to Fig. 5. a, Lifetimes of OT1 TCR bimolecular bonds with the indicated peptides bound to H2-Kbα3A2 or of the MHC–CD8 bond (wVSV) were measured by the BFP thermal fluctuation assay24 at zero-force. Their survival probabilities were plotted as fraction of events with a lifetime ≥ tb vs. lifetime tb. The number of bond lifetime measurements are: N = 52 (OVA), 39 (Q4), 49 (Q4R7), 26 (T4), 31 (Q4H7), 39 (Q7), 52 (G4), and 20 (VSV). b, Micrographs of the micropipette adhesion frequency assay25. A DP thymocyte (right) aspirated by a pipette was aligned with a RBC held by another pipette (left). The two cells were brought into a controlled contact for a given area (Ac) and duration (tc) and then retracted to detect binding, which was observed by the absence (top, no adhesion) or presence (bottom, adhesion) of RBC elongation. Scale bar = 5 µm. c, Specificity control. At 5 s contact, thymocytes adhered to RBCs coated with wOVA (13 μm-2) at higher frequency than those with mOVA (13 μm-2), showing the contribution of CD8 binding, but did not adhere to unmodified RBCs, biotinylated RBCs, and RBCs coated with biotin–SA (4335 μm-2) or mVSV (604 μm-2). Data are presented as mean ± s.e.m. of cell pairs each contacted 50 times to estimate an adhesion frequency. The number of cell pairs are: N = 3 (unmodified), 3 (biotin), 4 (biotin-SA), 3 (mVSV), 4 (mOVA), and 5 (wOVA). d, Adhesion frequency Pa vs. contact duration tc plots for indicated peptides bound to H2-Kbα3A2 (brown circle) or H2-Kb in the absence (blue square) or presence of anti-CD8 CT-CD8a (green triangle) or anti-TCR B20.1 (red diamond). Except for OVA whose measurements required a lower pMHC density (14-46 μm-2), a narrow range of pMHC densities (84-240 μm-2) were used to compare DP thymocyte adhesions in indicated conditions for each ligand. MHC–CD8 interaction (measured by using B20.1 to block TCR binding) mediated lower but clearly measureable adhesion frequencies than the total pMHC interactions with TCR and/or CD8. The much lower adhesions mediated by TCR than CD8 result from the 10 times lower expression of TCR than CD8 on DP thymocytes; but TCR still has higher affinities for pMHC than CD8 (cf. panel e). Since TCR–pMHC interactions (measured by using H2-Kbα3A2 or CT-CD8a blocking to abolish CD8 binding) were nearly undetectable for these peptides with such low pMHC densities, higher ligand densities (840-3739 μm-2) were used to measure their effective 2D affinities, as shown in panel e. Data are presented as mean ± s.e.m. of N≥3 cell pairs for 50 touches each pair. The data are representatives from at least two independent experiments per curve. e, No significant differences (p>0.5, one-way ANOVA) were observed among effective 2D affinities AcKa of TCR for pMHCs of distinctive selection outcomes (left ordinate, open bar, mean ± s.e.m., N≥5). MHC–CD8 AcKa (left ordinate, hatched bar, mean ± s.e.m., N=5) and available 3D tetramer values (right ordinate, black bar)4 are shown for comparison. f, To directly compare different ligands, the adhesion frequency data were converted to average number of bonds by <n> = – ln(1 – Pa), normalized by dividing them by the corresponding pMHC density mpMHC, and plotted versus contact duration tc. Except for OVA whose curve (red circle) is one log higher, the collapsed <ntotal>/mpMHC curves of the other ligands show no significant difference. The <nCD8>/mpMHC curve (black filled circle) is lower. g, Normalized total adhesion bonds, <ntotal>/mpMHC = – ln(1 – Pa)/mpMHC (open bar) for the indicated pMHCs (mean ± s.e.m., N≥3) and available 3D tetramer values (black bar)4 are shown for comparison. No significant differences were found among the middle 6 ligands (p>0.5, one-way ANOVA). The 2D kinetic parameters measured at zero force by adhesion frequency assay and thermal fluctuation assay are summarized in Supplementary Table 6
Supplementary Figure 4 Bond-lifetime distributions.
Related to Fig. 6. a,b, Survival probabilities of total OT1 TCR and/or CD8 bonds with the indicated peptides bound to H2-Kb (a) or bimolecular OT1 TCR bonds with the indicated peptides bound to H2-Kbα3A2 (b) measured by the force-clamp assay in the indicated force regimes. The MHC–CD8 interaction measured using VSV:H2-Kb is shown in black (a). c,d, Survival probabilities of total 2C TCR and/or CD8 bonds with the indicated peptides bound to H2-Kb or H-2Kbm3 (c) or bimolecular 2C TCR bonds with the indicated peptides bound to H2-Kbα3A2 (d) measured by the thermal fluctuation (0 pN) and force-clamp assay in the indicated force regimes. For the survival probabilities in (a-d), the higher the curve, the slower the dissociation, and the greater the number of bonds surviving a given time. The symbols and legend letters are color-coded to indicate thymocyte selection outcomes: red = negative selection, grey = selection threshold, and blue = positive selection. As force increases, the separation between red and blue curves increased, reached a maximum at 10-15 pN (third column), and then decreased. Separation in lifetime curves was greater for the total bonds that included the synergy between TCR and CD8 for pMHC binding (a, c) than bimolecular bonds between TCR and pMHC (b, d)
Supplementary Figure 5 Selection accuracy and ROC curves.
Related to Fig. 6. a, Selection accuracy SA versus threshold cumulative bond lifetime tth for a range of serial bond numbers n. b, SA versus n for a range tth at 0-5 pN with CD8 contribution, at 10-15 pN without CD8 contribution and at 0-5 pN without CD8 contribution. c, Receiver operating characteristic (ROC) curves plotting a thymocyte’s probability to be negatively selected by wQ4R7 (true-positive rate, or sensitivity) vs. its probability to be negatively selected by wQ4H7 (false-positive rate, or 1 - selectivity) for varying tth and a range of n. The three columns are calculated using the respective bond lifetime distributions of wQ4R7 and wQ4H7 at 0-5 pN, of mQ4R7 and mQ4H7 at 10-15 pN and of mQ4R7 and mQ4H7 at 0-5 pN from Supplementary Fig. 4a, b.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–6
Supplementary Video 1
Thymocytes exerting force on wQ4R7. Related to Figs. 3 and 4. Representative movie of a wild-type OT1 DP thymocyte pulling on wQ4R7. Thymocytes were placed on glass surfaces coated with wQ4R7 (tagged with a MTP with a force threshold of 13.1 pN) and anti-CD11a (not conjugated with any fluorescent dye). The de-quenched Cy5 signal indicates unfolding of the DNA hairpin by a >13.1 pN force generated by the thymocyte and applied to the pMHC via engaged TCR and CD8. Bright-field (left), Cy5 fluorescence (middle) and merged (right) images were recorded for 9 min.
Supplementary Video 2
Thymocytes exerting force on wQ4H7. Related to Figs. 3 and 4. Representative movie of a wild-type OT1 DP thymocyte pulling on wQ4H7. Thymocytes were placed on glass surfaces coated with wQ4H7 (tagged with a MTP with a force threshold of 13.1 pN) and anti-CD11a (not conjugated with any fluorescent dye). The de-quenched Cy5 signal indicates unfolding of the DNA hairpin by a >13.1 pN force generated by the thymocyte and applied to the pMHC via engaged TCR and CD8. Bright-field (left), Cy5 fluorescence (middle) and merged (right) images were recorded for 9 min.
Supplementary Video 3
3D images of a DP thymocytes pulling on wOVA. Related to Fig. 4. Rotating view of 3D reconstructed confocal images of a wild-type OT1 DP thymocyte placed on a glass surface, which was coated with wOVA (tagged with a MTP with a force threshold of 4.7 pN) and anti-CD11a (not conjugated with any fluorescent dye). The dequenched Cy5 signal indicates unfolding of the DNA hairpin by a >4.7 pN force generated by the thymocyte and applied to the pMHC via engaged TCR and CD8.
Rights and permissions
About this article
Cite this article
Hong, J., Ge, C., Jothikumar, P. et al. A TCR mechanotransduction signaling loop induces negative selection in the thymus. Nat Immunol 19, 1379–1390 (2018). https://doi.org/10.1038/s41590-018-0259-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-018-0259-z
This article is cited by
-
Unique roles of co-receptor-bound LCK in helper and cytotoxic T cells
Nature Immunology (2023)
-
Catch bond models may explain how force amplifies TCR signaling and antigen discrimination
Nature Communications (2023)
-
Cell volume controlled by LRRC8A-formed volume-regulated anion channels fine-tunes T cell activation and function
Nature Communications (2023)
-
Cooperative binding of T cell receptor and CD4 to peptide-MHC enhances antigen sensitivity
Nature Communications (2022)
-
Calculating the force-dependent unbinding rate of biological macromolecular bonds from force-ramp optical trapping assays
Scientific Reports (2022)