Abstract
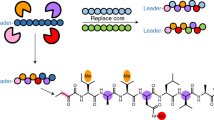
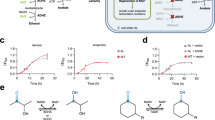
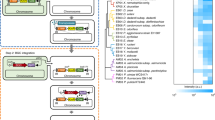
The modular nature of nonribosomal peptide biosynthesis has driven efforts to generate peptide analogs by substituting amino acid-specifying domains within nonribosomal peptide synthetase (NRPS) enzymes. Rational NRPS engineering has increasingly focused on finding evolutionarily favored recombination sites for domain substitution. Here we present an alternative evolution-inspired approach that involves large-scale diversification and screening. By amplifying amino acid-specifying domains en masse from soil metagenomic DNA, we substitute more than 1,000 unique domains into a pyoverdine NRPS. Initial fluorescence and mass spectrometry screens followed by sequencing reveal more than 100 functional domain substitutions, collectively yielding 16 distinct pyoverdines as major products. This metagenomic approach does not require the high success rates demanded by rational NRPS engineering but instead enables the exploration of large numbers of substitutions in parallel. This opens possibilities for the discovery and production of nonribosomal peptides with diverse biological activities.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available within the main text and its Supplementary Information file. Mass spectrometry data are available from https://doi.org/10.25345/C5S46HG94. The PDB file 6P1J is available from https://doi.org/10.2210/pdb6P1J/pdb. Source data are provided with this paper. Data are also available from the corresponding authors upon request.
Code availability
Code generated during this study is available from https://doi.org/10.5281/zenodo.8361572.
References
Newman, D. J. & Cragg, G. M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 83, 770–803 (2020).
Cook, M. A. & Wright, G. D. The past, present, and future of antibiotics. Sci. Transl. Med. 14, eabo7793 (2022).
Felnagle, E. A. et al. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol. Pharm. 5, 191–211 (2008).
Kunakom, S. & Eustaquio, A. S. Natural products and synthetic biology: where we are and where we need to go. mSystems 4, e00113-19 (2019).
Abbood, N., Präve, L., Bozhueyuek, K. A. J. & Bode, H. B. A practical guideline to engineering nonribosomal peptide synthetases. Methods Mol. Biol. 2670, 219–234 (2023).
WHO model list of essential medicines. World Health Organization www.who.int/publications/i/item/WHO-MHP-HPS-EML-2021.02 (2021).
Baltz, R. H. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 8, 557–563 (2008).
Bauman, K. D., Butler, K. S., Moore, B. S. & Chekan, J. R. Genome mining methods to discover bioactive natural products. Nat. Prod. Rep. 38, 2100–2129 (2021).
Milshteyn, A., Schneider, J. S. & Brady, S. F. Mining the metabiome: identifying novel natural products from microbial communities. Chem. Biol. 21, 1211–1223 (2014).
Hover, B. M. et al. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 3, 415–422 (2018).
Craig, J. W., Chang, F. Y., Kim, J. H., Obiajulu, S. C. & Brady, S. F. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl. Environ. Microbiol. 76, 1633–1641 (2010).
Brown, A. S., Calcott, M. J., Owen, J. G. & Ackerley, D. F. Structural, functional and evolutionary perspectives on effective re-engineering of non-ribosomal peptide synthetase assembly lines. Nat. Prod. Rep. 35, 1210–1228 (2018).
Winn, M., Fyans, J. K., Zhuo, Y. & Micklefield, J. Recent advances in engineering nonribosomal peptide assembly lines. Nat. Prod. Rep. 33, 317–347 (2016).
Baltz, R. H. Combinatorial biosynthesis of cyclic lipopeptide antibiotics: a model for synthetic biology to accelerate the evolution of secondary metabolite biosynthetic pathways. ACS Synth. Biol. 3, 748–758 (2014).
Calcott, M. J., Owen, J. G. & Ackerley, D. F. Efficient rational modification of non-ribosomal peptides by adenylation domain substitution. Nat. Commun. 11, 4554 (2020).
Fleischhacker, K. et al. De novo design and engineering of non-ribosomal peptide synthetases. Nat. Chem. 10, 275–281 (2018).
Bozhuyuk, K. A. J. et al. Modification and de novo design of non-ribosomal peptide synthetases using specific assembly points within condensation domains. Nat. Chem. 11, 653–661 (2019).
Bozhüyük, K. A. J. et al. Evolution inspired engineering of megasynthetases. Preprint at bioRxiv https://doi.org/10.1101/2022.12.02.518901 (2022).
Kries, H., Niquille, D. L. & Hilvert, D. A subdomain swap strategy for reengineering nonribosomal peptides. Chem. Biol. 22, 640–648 (2015).
Thong, W. L. et al. Gene editing enables rapid engineering of complex antibiotic assembly lines. Nat. Commun. 12, 6872 (2021).
Baunach, M., Chowdhury, S., Stallforth, P. & Dittmann, E. The landscape of recombination events that create nonribosomal peptide diversity. Mol. Biol. Evol. 38, 2116–2130 (2021).
Booth, T. J. et al. Bifurcation drives the evolution of assembly-line biosynthesis. Nat. Commun. 13, 3498 (2022).
Zhong, L. et al. Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis. Nat. Commun. 12, 296 (2021).
Charlop-Powers, Z. et al. Global biogeographic sampling of bacterial secondary metabolism. eLife 4, e05048 (2015).
Charlop-Powers, Z., Owen, J. G., Reddy, B. V. B., Ternei, M. A. & Brady, S. F. Chemical–biogeographic survey of secondary metabolism in soil. Proc. Natl Acad. Sci. USA 111, 3757–3762 (2014).
Liu, R., Li, X. & Lam, K. S. Combinatorial chemistry in drug discovery. Curr. Opin. Chem. Biol. 28, 117–126 (2017).
Marahiel, M. A., Stachelhaus, T. & Mootz, H. D. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97, 2651–2673 (1997).
Baltz, R. H. Natural product drug discovery in the genomic era: realities, conjectures, misconceptions, and opportunities. J. Ind. Microbiol. Biotechnol. 46, 281–299 (2019).
Schneider, T. D. & Stephens, R. M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990).
Voigt, C. A., Martinez, C., Wang, Z. G., Mayo, S. L. & Arnold, F. H. Protein building blocks preserved by recombination. Nat. Struct. Biol. 9, 553–558 (2002).
Tan, K. M. et al. Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains. Curr. Res. Struct. Biol. 2, 14–24 (2020).
Wang, Y. et al. ThreaDomEx: a unified platform for predicting continuous and discontinuous protein domains by multiple-threading and segment assembly. Nucleic Acids Res. 45, W400–W407 (2017).
Steiniger, C., Hoffmann, S. & Sussmuth, R. D. Desymmetrization of cyclodepsipeptides by assembly mode switching of iterative nonribosomal peptide synthetases. ACS Synth. Biol. 8, 661–667 (2019).
Yakimov, M. M., Giuliano, L., Timmis, K. N. & Golyshin, P. N. Recombinant acylheptapeptide lichenysin: high level of production by Bacillus subtilis cells. J. Mol. Microbiol. Biotechnol. 2, 217–224 (2000).
Doekel, S. et al. Non-ribosomal peptide synthetase module fusions to produce derivatives of daptomycin in Streptomyces roseosporus. Microbiology (Reading) 154, 2872–2880 (2008).
Mootz, H. D., Schwarzer, D. & Marahiel, M. A. Construction of hybrid peptide synthetases by module and domain fusions. Proc. Natl Acad. Sci. USA 97, 5848–5853 (2000).
Linhart, C. & Shamir, R. The degenerate primer design problem: theory and applications. J. Comput. Biol. 12, 431–456 (2005).
Hsieh, T. C., Ma, K. H. & Chao, A. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 7, 1451–1456 (2016).
Calcott, M. J., Owen, J. G., Lamont, I. L. & Ackerley, D. F. Biosynthesis of novel pyoverdines by domain substitution in a nonribosomal peptide synthetase of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 80, 5723–5731 (2014).
Rehm, K., Vollenweider, V., Kümmerli, R. & Bigler, L. A comprehensive method to elucidate pyoverdines produced by fluorescent Pseudomonas spp. by UHPLC-HR-MS/MS. Anal. Bioanal. Chem. 414, 2671–2685 (2022).
Meyer, J.-M. et al. Siderotyping of fluorescent Pseudomonas: molecular mass determination by mass spectrometry as a powerful pyoverdine siderotyping method. Biometals 21, 259–271 (2008).
Reetz, M. T. & Carballeira, J. D. Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat. Protoc. 2, 891–903 (2007).
Acevedo-Rocha, C. G., Hoebenreich, S. & Reetz, M. T. Iterative saturation mutagenesis: a powerful approach to engineer proteins by systematically simulating Darwinian evolution. Methods Mol. Biol. 1179, 103–128 (2014).
Yang, J. Y. et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 76, 1686–1699 (2013).
Owen, J. G., Calcott, M. J., Robins, K. J. & Ackerley, D. F. Generating functional recombinant NRPS enzymes in the laboratory setting via peptidyl carrier protein engineering. Cell Chem. Biol. 23, 1395–1406 (2016).
Evans, B. S., Chen, Y. Q., Metcalf, W. W., Zhao, H. M. & Kelleher, N. L. Directed evolution of the nonribosomal peptide synthetase AdmK generates new andrimid derivatives in vivo. Chem. Biol. 18, 601–607 (2011).
Azuma, T., Harrison, G. I. & Demain, A. L. Isolation of a gramicidin S hyperproducing strain of Bacillus brevis by use of a flourescence activated cell sorting system. Appl. Microbiol. Biotechnol. 38, 173–178 (1992).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Abdallah, M. A. & Meyer, J. M. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. Microbiology 107, 319–328 (1978).
Xiao, R. & Kisaalita, W. S. Purification of pyoverdines of Pseudomonas fluorescens 2-79 by copper chelate chromatography. Appl. Environ. Microbiol. 61, 3769–3774 (1995).
Stevenson, L. J., Ackerley, D. F. & Owen, J. G. Preparation of soil metagenome libraries and screening for gene-specific amplicons. Methods Mol. Biol. 2397, 3–17 (2022).
Schmid, R. et al. Integrative analysis of multimodal mass spectrometry data in MZmine 3. Nat. Biotechnol. 41, 447–449 (2023).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Ono, K., Muetze, T., Kolishovski, G., Shannon, P. & Demchak, B. CyREST: turbocharging cytoscape access for external tools via a RESTful API. F1000Res. 4, 478 (2015).
Cornelis, P. Unexpected interaction of a siderophore with aluminum and its receptor. J. Bacteriol. 190, 6541–6543 (2008).
Acknowledgements
This work was supported by the Royal Society of New Zealand Marsden Fund (grant no. 18-VUW-082 to M.J.C. and G.L.C.), and the Health Research Council of New Zealand (contract 16/172 to D.F.A. and J.G.O.). S.R.M. was supported by a New Zealand Lottery Health Research Scholarship (contract LHR-2020-129624). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
S.R.M., D.F.A. and M.J.C. conceived and designed the work. S.R.M., E.M.R.M. and M.J.C. performed the experiments. S.R.M. and M.J.C. performed the data analysis and prepared all figures. L.J.S., J.G.O. and M.J.C. created the SL and RX metagenomic libraries. M.J.C., G.L.C., D.F.A. and J.G.O. obtained funding for this study. S.R.M., D.F.A. and M.J.C. wrote the manuscript. S.R.M., G.L.C., J.G.O., D.F.A. and M.J.C. edited the manuscript, and all authors approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Donatella de Pascale, Sergey Zotchev and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11 and Tables 1–4.
Supplementary Data 1
Raw data for Supplementary Fig. 8.
Supplementary Data 2
DNA sequences and oligonucleotides.
Source data
Source Data Fig. 3
Raw data.
Source Data Fig. 4
Raw data.
Source Data Fig. 5
Raw data.
Source Data Fig. 6
Raw data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Messenger, S.R., McGuinniety, E.M.R., Stevenson, L.J. et al. Metagenomic domain substitution for the high-throughput modification of nonribosomal peptides. Nat Chem Biol 20, 251–260 (2024). https://doi.org/10.1038/s41589-023-01485-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-023-01485-1