Abstract
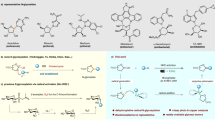
The anthraquinone-fused enediynes (AFEs) combine an anthraquinone moiety and a ten-membered enediyne core capable of generating a cytotoxic diradical species. AFE cyclization is triggered by opening the F-ring epoxide, which is also the site of the most structural diversity. Previous studies of tiancimycin A, a heavily modified AFE, have revealed a cryptic aldehyde blocking installation of the epoxide, and no unassigned oxidases could be predicted within the tnm biosynthetic gene cluster. Here we identify two consecutively acting cofactorless oxygenases derived from methyltransferase and α/β-hydrolase protein folds, TnmJ and TnmK2, respectively, that are responsible for F-ring tailoring in tiancimycin biosynthesis by comparative genomics. Further biochemical and structural characterizations reveal that the electron-rich AFE anthraquinone moiety assists in catalyzing deformylation, epoxidation and oxidative ring cleavage without exogenous cofactors. These enzymes therefore fill important knowledge gaps for the biosynthesis of this class of molecules and the underappreciated family of cofactorless oxygenases.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available within the main text and its Supplementary Information file. Data are available from the corresponding author upon request. The coordinates of the TnmJ, TnmK2 and TnmK2–9 structures have been deposited to the PDB with the accession codes 8G5S, 8G5T and 8G5U, respectively. Source data are provided with this paper.
References
Adhikari A., Teijaro C. N., Townsend C. A. & Shen B. in Comprehensive Natural Products III (eds Liu, H.-W. & Begley, T. P.) 365–414 (Elsevier, 2020).
Adhikari, A., Shen, B. & Rader, C. Challenges and opportunities to develop enediyne natural products as payloads for antibody-drug conjugates. Antib. Ther. 4, 1–15 (2021).
Maeda, H. SMANCS and polymer-conjugated macromolecular drugs: advantages in cancer chemotherapy. Adv. Drug Deliv. Rev. 46, 169–185 (2001).
Konishi, M. et al. Dynemicin A, a novel antibiotic with the anthraquinone and 1,5-diyn-3-ene subunit. J. Antibiot. 42, 1449–1452 (1989).
Davies, J. et al. Uncialamycin, a new enediyne antibiotic. Org. Lett. 7, 5233–5236 (2005).
Yan, X. et al. Strain prioritization and genome mining for enediyne natural products. mBio 7, e02104–e02116 (2016).
Nicolaou, K. C. et al. Uncialamycin-based antibody–drug conjugates: unique enediyne ADCs exhibiting bystander killing effect. Proc. Natl Acad. Sci. USA 118, e2107042118 (2021).
Low, Z. J. et al. Sungeidines from a non-canonical enediyne biosynthetic pathway. J. Am. Chem. Soc. 142, 1673–1679 (2020).
Steele, A. D. et al. Application of a biocatalytic strategy for the preparation of tiancimycin-based antibody–drug conjugates revealing key insights into structure–activity relationships. J. Med. Chem. 66, 1562–1573 (2023).
Igarashi, M. et al. Sealutomicins, new enediyne antibiotics from the deep-sea actinomycete Nonomuraea sp. MM565M-173N2. J. Antibiot. 74, 291–299 (2021).
Gui, C. et al. Intramolecular C–C bond formation links anthraquinone and enediyne scaffolds in tiancimycin biosynthesis. J. Am. Chem. Soc. 144, 20452–20462 (2022).
Yan, X. et al. Comparative studies of the biosynthetic gene clusters for anthraquinone-fused enediynes shedding light into the tailoring steps of tiancimycin biosynthesis. Org. Lett. 20, 5918–5921 (2018).
Machovina, M. M., Usselman, R. J. & DuBois, J. L. Monooxygenase substrates mimic flavin to catalyze cofactorless oxygenations. J. Biol. Chem. 291, 17816–17828 (2016).
Thierbach, S. et al. Substrate-assisted O2 activation in a cofactor-independent dioxygenase. Chem. Biol. 21, 217–225 (2014).
Hernández-Ortega, A. et al. Catalytic mechanism of cofactor-free dioxygenases and how they circumvent spin-forbidden oxygenation of their substrates. J. Am. Chem. Soc. 137, 7474–7487 (2015).
Machovina, M. M., Ellis, E. S., Carney, T. J., Brushett, F. R. & DuBois, J. L. How a cofactor-free protein environment lowers the barrier to O2 reactivity. J. Biol. Chem. 294, 3661–3669 (2019).
Steiner, R. A., Janssen, H. J., Roversi, P., Oakley, A. J. & Fetzner, S. Structural basis for cofactor-independent dioxygenation of N-heteroaromatic compounds at the α/β-hydrolase fold. Proc. Natl Acad. Sci. USA 107, 657–662 (2010).
Jansson, A. et al. Aclacinomycin 10-hydroxylase is a novel substrate-assisted hydroxylase requiring S-adenosyl-l-methionine as cofactor. J. Biol. Chem. 280, 3636–3644 (2005).
Fetzner, S. & Steiner, R. A. Cofactor-independent oxidases and oxygenases. Appl. Microbiol. Biotechnol. 86, 791–804 (2010).
Matthews, J. C., Hori, K. & Cormier, M. J. Substrate and substrate analog binding properties of Renilla luciferase. Biochemistry 16, 5217–5220 (1977).
Sarma, A. D. & Tipton, P. A. Evidence for urate hydroperoxide as an intermediate in the urate oxidase reaction. J. Am. Chem. Soc. 122, 11252–11253 (2000).
Sciara, G. et al. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO J. 22, 205–215 (2003).
Widboom, P. F., Fielding, E. N., Liu, Y. & Bruner, S. D. Structural basis for cofactor-independent dioxygenation in vancomycin biosynthesis. Nature 447, 342–345 (2007).
Siitonen, V., Blauenburg, B., Kallio, P., Mäntsälä, P. & Metsä-Ketelä, M. Discovery of a two-component monooxygenase SnoaW/SnoaL2 involved in nogalamycin biosynthesis. Chem. Biol. 19, 638–646 (2012).
Shen, B. & Hutchinson, C. R. Tetracenomycin F1 monooxygenase: oxidation of a naphthacenone to a naphthacenequinone in the biosynthesis of tetracenomycin C in Streptomyces glaucescens. Biochemistry 32, 6656–6663 (1993).
Tokiwa, Y. et al. Biosynthesis of dynemicin A, a 3-ene-1,5-diyne antitumor antibiotic. J. Am. Chem. Soc. 114, 4107–4110 (1992).
Nicolaou, K. C. et al. Total synthesis and biological evaluation of tiancimycins A and B, yangpumicin A, and related anthraquinone-fused enediyne antitumor antibiotics. J. Am. Chem. Soc. 142, 2549–2561 (2020).
Bar-Even, A. et al. The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 50, 4402–4410 (2011).
Marsh, E. N. G. & Waugh, M. W. Aldehyde decarbonylases: enigmatic enzymes of hydrocarbon biosynthesis. ACS Catal. 3, 2515–2521 (2013).
Qiu, Y. et al. An insect-specific P450 oxidative decarbonylase for cuticular hydrocarbon biosynthesis. Proc. Natl Acad. Sci. USA 109, 14858–14863 (2012).
Schirmer, A., Rude, M. A., Li, X., Popova, E. & del Cardayre, S. B. Microbial biosynthesis of alkanes. Science 329, 559–562 (2010).
Warui, D. M. et al. Detection of formate, rather than carbon monoxide, as the stoichiometric coproduct in conversion of fatty aldehydes to alkanes by a cyanobacterial aldehyde decarbonylase. J. Am. Chem. Soc. 133, 3316–3319 (2011).
Schneider-Belhaddad, F. & Kolattukudy, P. Solubilization, partial purification, and characterization of a fatty aldehyde decarbonylase from a higher plant, Pisum sativum. Arch. Biochem. Biophys. 377, 341–349 (2000).
Yoshimoto, F. K. & Guengerich, F. P. Mechanism of the third oxidative step in the conversion of androgens to estrogens by cytochrome P450 19A1 steroid aromatase. J. Am. Chem. Soc. 136, 15016–15025 (2014).
He, H.-Y. & Ryan, K. S. Glycine-derived nitronates bifurcate to O-methylation or denitrification in bacteria. Nat. Chem. 13, 599–606 (2021).
Jonnalagadda, R. et al. Biochemical and crystallographic investigations into isonitrile formation by a nonheme iron-dependent oxidase/decarboxylase. J. Biol. Chem. 296, 100231 (2021).
Zhang, Z. et al. Enzyme-catalyzed inverse-electron demand Diels–Alder reaction in the biosynthesis of antifungal ilicicolin H. J. Am. Chem. Soc. 141, 5659–5663 (2019).
Townsend, C. A. New reactions in clavulanic acid biosynthesis. Curr. Opin. Chem. Biol. 6, 583–589 (2002).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Gust, B., Challis, G. L., Fowler, K., Kieser, T. & Chater, K. F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl Acad. Sci. USA 100, 1541–1546 (2003).
Acknowledgements
This work was supported in part by National Institutes of Health (NIH) grants GM134954 (B.S.) and OD021550 (NMR Core Facility). This work was supported in part by NIH postdoctoral fellowships GM134688 (E.K.) and GM133114 (A.D.S.). We thank X. Kong of the NMR Core Facility at The Herbert Wertheim UF Scripps Institute for Biomedical Innovation & Technology, University of Florida, Jupiter, Florida, for assistance with NMR analysis.
Author information
Authors and Affiliations
Contributions
C.G., E.K., Y.-C.L., G.L., A.D.S., D.Y. and C.C. performed experiments and analysis. C.G., E.K. and B.S. conceived of the project and designed experiments. C.G., E.K. and B.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Jennifer DuBois, Jarrod French and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Evaluation of TnmJ for metal, SAM, and reducing agent dependence.
a, No difference was observed upon addition of 10 mM EDTA to a reaction with TnmJ and 8. b, No difference was observed upon addition of 2 mM reducing agents DTT or TCEP. c, No difference was observed upon addition of 200 μM SAM or SAH to a reaction with TnmJ and 8. Likewise, charcoal-treated TnmJ showed no difference. Bars depict the mean; error bars represent standard deviation (n = 3 independent experiments).
Extended Data Fig. 2 Mutagenesis of TnmJ.
Site-directed mutants of TnmJ were evaluated in vitro with 100 μM 8. Results were used to evaluate possible binding modes from molecular dynamics simulations. Bars depict the mean; error bars represent standard deviation (n = 3 independent experiments).
Extended Data Fig. 3 Mutagenesis of TnmK2.
TnmK2 mutants were evaluated in vitro with 13 produced in situ by TnmJ from 100 μM 8. N.D. = not detected. Bars depict the mean; error bars represent standard deviation (n = 3 independent experiments).
Supplementary information
Supplementary Information
Supplementary methods, Figs. 1–44 and Tables 1–10.
Supplementary Data
Statistical source data for Supplementary Figs. 20, 27, 31, 33 and 43.
Source data
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gui, C., Kalkreuter, E., Liu, YC. et al. Cofactorless oxygenases guide anthraquinone-fused enediyne biosynthesis. Nat Chem Biol 20, 243–250 (2024). https://doi.org/10.1038/s41589-023-01476-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-023-01476-2