Abstract
Bio-refining lignocellulose could provide a sustainable supply of fuels and fine chemicals; however, the challenges associated with the co-utilization of xylose and glucose typically compromise the efficiency of lignocellulose conversion. Here we engineered the industrial yeast Ogataea polymorpha (Hansenula polymorpha) for lignocellulose biorefinery by facilitating the co-utilization of glucose and xylose to optimize the production of free fatty acids (FFAs) and 3-hydroxypropionic acid (3-HP) from lignocellulose. We rewired the central metabolism for the enhanced supply of acetyl-coenzyme A and nicotinamide adenine dinucleotide phosphate hydrogen, obtaining 30.0 g l−1 of FFAs from glucose, with productivity of up to 0.27 g l−1 h−1. Strengthening xylose uptake and catabolism promoted the synchronous utilization of glucose and xylose, which enabled the production of 38.2 g l−1 and 7.0 g l−1 FFAs from the glucose–xylose mixture and lignocellulosic hydrolysates, respectively. Finally, this efficient cell factory was metabolically transformed for 3-HP production with the highest titer of 79.6 g l−1 in fed-batch fermentation in mixed glucose and xylose.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available within the article, Supplementary Information, and source data files linked to each figure. Source data are provided with this paper.
Code availability
No custom code was used in this paper.
References
Uthandi, S. et al. Microbial biodiesel production from lignocellulosic biomass: new insights and future challenges. Crit. Rev. Environ. Sci. Technol. 52, 2197–2225 (2022).
Chandel, A. K., Garlapati, V. K., Singh, A. K., Antunes, F. A. F. & da Silva, S. S. The path forward for lignocellulose biorefineries: bottlenecks, solutions, and perspective on commercialization. Bioresour. Technol. 264, 370–381 (2018).
Reshmy, R. et al. Lignocellulose in future biorefineries: strategies for cost-effective production of biomaterials and bioenergy. Bioresour. Technol. 344, 126241 (2022).
Usmani, Z. et al. Lignocellulosic biorefineries: the current state of challenges and strategies for efficient commercialization. Renew. Sustain. Energy Rev. 148, 111258 (2021).
Zhang, G.-C., Liu, J.-J., Kong, I. I., Kwak, S. & Jin, Y.-S. Combining C6 and C5 sugar metabolism for enhancing microbial bioconversion. Curr. Opin. Chem. Biol. 29, 49–57 (2015).
Sun, L. & Jin, Y.-S. Xylose assimilation for the efficient production of biofuels and chemicals by engineered Saccharomyces cerevisiae. Biotechnol. J. 16, 2000142 (2021).
Menon, V. & Rao, M. Trends in bioconversion of lignocellulose: biofuels, platform chemicals & biorefinery concept. Prog. Energy Combust. Sci. 38, 522–550 (2012).
Kim, S. R., Ha, S.-J., Wei, N., Oh, E. J. & Jin, Y.-S. Simultaneous co-fermentation of mixed sugars: a promising strategy for producing cellulosic ethanol. Trends Biotechnol. 30, 274–282 (2012).
Kwak, S., Jo, J. H., Yun, E. J., Jin, Y.-S. & Seo, J.-H. Production of biofuels and chemicals from xylose using native and engineered yeast strains. Biotechnol. Adv. 37, 271–283 (2019).
Zhou, H., Cheng, J.-S., Wang, B. L., Fink, G. R. & Stephanopoulos, G. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab. Eng. 14, 611–622 (2012).
Young, E. M., Comer, A. D., Huang, H. & Alper, H. S. A molecular transporter engineering approach to improving xylose catabolism in Saccharomyces cerevisiae. Metab. Eng. 14, 401–411 (2012).
Young, E. M., Tong, A., Bui, H., Spofford, C. & Alper, H. S. Rewiring yeast sugar transporter preference through modifying a conserved protein motif. Proc. Natl Acad. Sci. USA 111, 131–136 (2014).
Ha, S.-J. et al. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc. Natl Acad. Sci. USA 108, 504–509 (2011).
Kurylenko, O. O. et al. Peroxisomes and peroxisomal transketolase and transaldolase enzymes are essential for xylose alcoholic fermentation by the methylotrophic thermotolerant yeast, Ogataea (Hansenula) polymorpha. Biotechnol. Biofuels 11, 16 (2018).
Ryabova, O. B., Chmil, O. M. & Sibirny, A. A. Xylose and cellobiose fermentation to ethanol by the thermotolerant methylotrophic yeast Hansenula polymorpha. FEMS Yeast Res. 4, 157–164 (2003).
Saraya, R. et al. Novel genetic tools for Hansenula polymorpha. FEMS Yeast Res. 12, 271–278 (2012).
Ubiyvovk, V. M., Ananin, V. M., Malyshev, A. Y., Kang, H. A. & Sibirny, A. A. Optimization of glutathione production in batch and fed-batch cultures by the wild-type and recombinant strains of the methylotrophic yeast Hansenula polymorpha DL-1. BMC Biotechnol. 11, 8 (2011).
Ruchala, J., Kurylenko, O. O., Dmytruk, K. V. & Sibirny, A. A. Construction of advanced producers of first- and second-generation ethanol in Saccharomyces cerevisiae and selected species of non-conventional yeasts (Scheffersomyces stipitis, Ogataea polymorpha). J. Ind. Microbiol. Biotechnol. 47, 109–132 (2020).
Voronovsky, A. Y., Rohulya, O. V., Abbas, C. A. & Sibirny, A. A. Development of strains of the thermotolerant yeast Hansenula polymorpha capable of alcoholic fermentation of starch and xylan. Metab. Eng. 11, 234–242 (2009).
Li, Y. et al. Production of free fatty acids from various carbon sources by Ogataea polymorpha. Bioresour. Bioprocess. 9, 78 (2022).
Thorwall, S., Schwartz, C., Chartron, J. W. & Wheeldon, I. Stress-tolerant non-conventional microbes enable next-generation chemical biosynthesis. Nat. Chem. Biol. 16, 113–121 (2020).
Peralta-Yahya, P. P., Zhang, F., Del Cardayre, S. B. & Keasling, J. D. Microbial engineering for the production of advanced biofuels. Nature 488, 320–328 (2012).
Choi, S., Song, C. W., Shin, J. H. & Lee, S. Y. Biorefineries for the production of top building block chemicals and their derivatives. Metab. Eng. 28, 223–239 (2015).
Gao, J., Li, Y., Yu, W. & Zhou, Y. J. Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol. Nat. Metab. 4, 932–943 (2022).
Blazeck, J. et al. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 5, 3131 (2014).
Xue, Z. et al. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat. Biotechnol. 31, 734–740 (2013).
Bocanegra, J. A., Scrutton, N. S. & Perham, R. N. Creation of an NADP-dependent pyruvate dehydrogenase multienzyme complex by protein engineering. Biochemistry 32, 2737–2740 (1993).
Feng, D., Gao, J., Gong, Z. & Zhou, Y. J. Production of fatty acids by engineered Ogataea polymorpha. Chin. J. Biotechnol. 38, 760–771 (2022).
Charoenrat, T., Antimanon, S., Kocharin, K., Tanapongpipat, S. & Roongsawang, N. High cell density process for constitutive production of a recombinant phytase in thermotolerant methylotrophic yeast Ogataea thermomethanolica using table sugar as carbon source. Appl. Biochem. Biotechnol. 180, 1618–1634 (2016).
Dmytruk, O. V., Dmytruk, K. V., Abbas, C. A., Voronovsky, A. Y. & Sibirny, A. A. Engineering of xylose reductase and overexpression of xylitol dehydrogenase and xylulokinase improves xylose alcoholic fermentation in the thermotolerant yeast Hansenula polymorpha. Microb. Cell Fact. 7, 21 (2008).
Lee, M., Rozeboom, H. J., Keuning, E., Waal, P. D. & Janssen, D. B. Structure-based directed evolution improves S. cerevisiae growth on xylose by influencing in vivo enzyme performance. Biotechnol. Biofuels 13, 5 (2020).
Seike, T., Kobayashi, Y., Sahara, T., Ohgiya, S. & Fujimori, K. E. Molecular evolutionary engineering of xylose isomerase to improve its catalytic activity and performance of micro-aerobic glucose/xylose co-fermentation in Saccharomyces cerevisiae. Biotechnol. Biofuels 12, 139 (2019).
Vasylyshyn, R. et al. Engineering of sugar transporters for improvement of xylose utilization during high-temperature alcoholic fermentation in Ogataea polymorpha yeast. Microb. Cell Fact. 19, 96 (2020).
Krahulec, S. et al. Fermentation of mixed glucose-xylose substrates by engineered strains of Saccharomyces cerevisiae: role of the coenzyme specificity of xylose reductase, and effect of glucose on xylose utilization. Microb. Cell Fact. 9, 16 (2010).
Voronovsky, A. Y. et al. Expression of xylA genes encoding xylose isomerases from Escherichia coli and Streptomyces coelicolor in the methylotrophic yeast Hansenula polymorpha. FEMS Yeast Res. 5, 1055–1062 (2005).
Papapetridis, I. et al. Laboratory evolution for forced glucose-xylose co-consumption enables identification of mutations that improve mixed-sugar fermentation by xylose-fermenting Saccharomyces cerevisiae. FEMS Yeast Res. 18, foy056 (2018).
Matsushika, A., Nagashima, A., Goshima, T. & Hoshino, T. Fermentation of xylose causes inefficient metabolic state due to carbon/energy starvation and reduced glycolytic flux in recombinant industrial Saccharomyces cerevisiae. PLoS ONE 8, e69005 (2013).
Jin, Y.-S., Laplaza, J. M. & Jeffries, T. W. Saccharomyces cerevisiae engineered for xylose metabolism exhibits a respiratory response. Appl. Environ. Microbiol. 70, 6816–6825 (2004).
Xu, J. Z., Zhang, J. L., Guo, Y. F., Jia, Q. D. & Zhang, W. G. Heterologous expression of Escherichia coli fructose-1,6-bisphosphatase in Corynebacterium glutamicum and evaluating the effect on cell growth and l-lysine production. Prep. Biochem. Biotechnol. 44, 493–509 (2014).
Georgi, T., Rittmann, D. & Wendisch, V. F. Lysine and glutamate production by Corynebacterium glutamicum on glucose, fructose and sucrose: roles of malic enzyme and fructose-1,6-bisphosphatase. Metab. Eng. 7, 291–301 (2005).
Becker, J. et al. Metabolic flux engineering of L-lysine production in Corynebacterium glutamicum–over expression and modification of G6P dehydrogenase. J. Biotechnol. 132, 99–109 (2007).
Sumita, T. et al. Peroxisome deficiency represses the expression of n-alkane-inducible YlALK1 encoding cytochrome P450ALK1 in Yarrowia lipolytica. FEMS Microbiol. Lett. 214, 31–38 (2002).
Yu, T. et al. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell 174, 1549–1558 (2018).
Zhou, Y. J. et al. Harnessing yeast peroxisomes for biosynthesis of fatty-acid-derived biofuels and chemicals with relieved side-pathway competition. J. Am. Chem. Soc. 138, 15368–15377 (2016).
Gao, J., Gao, N., Zhai, X. & Zhou, Y. J. Recombination machinery engineering for precise genome editing in methylotrophic yeast Ogataea polymorpha. iScience 24, 102168 (2021).
Yu, W., Gao, J., Zhai, X. & Zhou, Y. J. Screening neutral sites for metabolic engineering of methylotrophic yeast Ogataea polymorpha. Synth. Syst. Biotechnol. 6, 63–68 (2021).
Chen, X. et al. Densifying lignocellulosic biomass with alkaline chemicals (DLC) pretreatment unlocks highly fermentable sugars for bioethanol production from corn stover. Green Chem. 23, 4828–4839 (2021).
Zhou, Y. J. et al. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat. Commun. 7, 11709 (2016).
Acknowledgements
We would like to thank the Energy Biotechnology Platform of DICP for providing facility assistance. This study was financially supported by the National Natural Science Foundation of China (22161142008 and 21922812) and a DICP innovation grant (DICP I202210) from the Dalian Institute of Chemical Physics, Chinese Academy of Sciences (CAS).
Author information
Authors and Affiliations
Contributions
J.G. and Y.J.Z. conceived the study. J.G. and W.Y. designed and performed most of the experiments. Y.L. performed fermentation experiments. M.J. prepared lignocellulosic hydrolysates. J.G., L.Y. and Y.J.Z. wrote, reviewed and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
A patent application (application no. 202310229165.5), for protecting the production of free fatty acids and 3-hydroxypropionic acid from glucose and xylose in O. polymorpha, has been filed by the Dalian Institute of Chemical Physics, Chinese Academy of Sciences, with Y.J.Z., J.G. and W.Y. named as inventors.
Peer review
Peer review information
Nature Chemical Biology thanks Bing-Zhi Li, Tsutomu Tanaka and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Metabolic rewiring for enhanced FFA production in O. polymorpha.
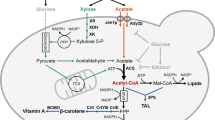
FA production requires enhanced supply of cofactor NADPH and precursor acetyl-CoA. To promote NADPH supply, pentose phosphate pathway (ZWF1) and gluconeogenesis (FBP1 and PCK1) are adopted. To promote acetyl-CoA supply, three independent strategies, including a modifiedβ-oxidation (pex10Δ), citrate lysis (MmACL, ScYHM2, AMPD), and PDH mutant (PDHm) were adopted, coupled with the overexpression of gene ACC1 to enhance malonyl-CoA supply.
Extended Data Fig. 2 Fluorescence subcellular localization of peroxisomes in control strain and pex10Δ strain.
GFP with the signal peptide SKL was targeted to peroxisomes in control strain and pex10Δ strain. Engineered strains were cultivated in minimal medium with 20 g/L glucose for 48 h, and cells were collected. GFP fluorescence in control strain was much focused, demonstrating the complete and functional peroxisomes. In contrast, GFP fluorescence in pex10Δ strain was partially dispersed into cytosol, demonstrating the destroyed peroxisome biogenesis for the enhanced supply of acetyl-CoA and acyl-CoA. The data result from at least two independent experiments with at least hree replicates in one experiment.
Extended Data Fig. 3 Cell growth and FFA production by the introduction of PDH complex and citrate lysis pathway.
a, The overexpressed PDH complex decreased FFA titers. Genes of pyruvate dehydrogenase (PDH) complex from E.coli, including a NADP+-dependent pyruvate dehydrogenase variant (EcLpdA*), dihydrolipoamide transacetylase (EcAceE), and dihydrolipoamide dehydrogenase (EcAceF), were introduced into O. polymorpha. b, Sketch map of citrate lysis pathway. Citrate was lysed into acetyl-CoA by a heterogeneous ATP citrate lyase from Mus musculus (MmACL), and a specific mitochondrial citrate transporter from S. cerevisiae (ScYHM2), and AMP deaminase (AMPD) to promote the accumulation of cytoplasmic citrate. c, Effects of overexpression of genes from citrate lysis on cell growth and FFA production. Removing the over-expressed gene AMPD in strain HpFA27L and HpFA42L had no significant effects on both FFA production (d) and cell growth (e). f, Over-expression of gene ScYHM2 partially hindered cell growth and decreased FFA production. The data are presented as the mean ± s.e.m. (n = 3 biologically independent samples).
Extended Data Fig. 4 Overexpression of MmACL and ScIDP2 in strain HpFA40 failed to promote both cell growth and FFA production.
The data are presented as the mean ± s.e.m. (n = 3 biologically independent samples).
Extended Data Fig. 5 Overexpression of gene ACC1 severely hindered cell growth.
An enhanced ACC1 expression by replacing its own promoter with PGAP in all engineered strains hindered cell growth (a), and decreased FFA titer (b). c, Overexpression of gene ACC1 by either PGAP or PTEF1 repressed cell growth, which indicated gene ACC1 was strictly regulated in O. polymorpha. The data are presented as the mean ± s.e.m. (n = 3 biologically independent samples).
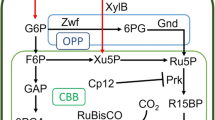
Extended Data Fig. 6 Strengthened xylose assimilation for over-production of FFAs in O. polymorpha.
To strengthen xylose assimilation, xylose isomerases (XI) from both Piromyces Sp. (PspXI*) and Lachnoclostridium phytofermentans (LpXI*), coupled with native xylulokinase (XYL3), were introduced. Engineered strains were cultivated in minimal medium containing 10 g/L xylose, and cell growth (a), xylose utilization (b), and FFA production (c) were detected. d, Xylitol accumulation in control strain HpFA10 and engineered strain HpFA50 when cultivated in sole xylose within 132 h. e, Cofactor quantification in control strain and engineered strains when cultivated in sole xylose. Strains were cultivated at 37 °C, 220 rpm for 96 h, and data are presented as mean ± s.e.m. (n = 3 biologically independent samples).
Extended Data Fig. 7 Synchronous utilization of xylose and glucose for FFA production in O. polymorpha.
A hexose transporter mutant with a higher affinity to xylose (HXT1*) was introduced to strain HpFA50 under control of PGAP (HpFA53) and PPGD1 (HpFA54). On this basis, FBP1-ZWF1 were over-expressed in strain HpFA50, HpFA53, and HpFA54, generating strain HpFA55, HpFA56, and HpFA57, respectively, to enhance the supply of NADPH. All engineered strains were cultured in 10 g/L xylose (X10) to measure the cell growth (a) and xylose utilization (b). FFA titers were measured after 120 h of cultivation at 37 °C, 220 rpm (c). On the other hand, a mixture of 20 g/L glucose and 10 g/L xylose (G20X10) was adopted to conduct the batch fermentation in flasks, demonstrating cell growth (d), and the synchronous utilization of glucose (e) and xylose (f). Data are presented as mean ± s.e.m. (n = 3 biologically independent samples).
Extended Data Fig. 8 Cell growth and sugar utilization of strain HpFA58 in glucose, or/and xylose.
To promote fermentative performances, gene KU80, URA3, and LEU2 were supplemented into strain HpFA56, generating strain HpFA58. Fermentations were carried out in 20 g/L glucose (a), 10 g/L xylose (b), 20 g/L glucose+10 g/L xylose (c), to measure cell growth and xylose utilization. Strains were cultivated at 37 °C, 220 rpm for 72 h, and data are presented as mean ± s.e.m. (n = 3 biologically independent samples).
Extended Data Fig. 9 Metabolic transforming of O. polymorpha from FFA to 3-HP.
a, To recover FAA1 gene, its native terminator was replaced by TADH1 by using CRISPR-Cas9 technique. Subsequently, gene MCR under control of promoter PGAP was integrated into 4NS7 locus. PFAA1: FAA1 promoter; TFAA1: FAA1 terminator; TADH1: ADH1 terminator; FAA1dw: downstream homologous arm of FAA1 terminator; PGAP: GAP promoter; TDIT1: DIT1 terminator; 4NS7up: upstream homologous arm of 4NS7 locus; 4NS7dw: downstream homologous arm of 4NS7 locus. b, 3-HP was produced in strain XFM and GFM from 20 g/L xylose and 20 g/L glucose in minimal medium, respectively, after 72 h of cultivation at 37 °C, 220 rpm. Data are presented as mean ± stand deviation. (n = 3 biologically independent samples).
Extended Data Fig. 10 Flowchart of O. polymorpha strain construction for FFA and 3-HP production.
O. polymorpha with a disrupted gene faa1Δ was adopted for FFA accumulation, and the supply of acetyl-CoA and NADPH was enhanced for FFA overproduction. Subsequently, the xylose uptake and catabolism were enhanced for the simultaneous co-utilization of glucose and xylose. Metabolic transforming strategy was introduced to convert the FFA producing strain to 3-HP production. Gene in blue indicated the specific genetic manipulation during the metabolic engineering route (red arrow).
Supplementary information
Supplementary Information
Supplementary Tables 1–7.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gao, J., Yu, W., Li, Y. et al. Engineering co-utilization of glucose and xylose for chemical overproduction from lignocellulose. Nat Chem Biol 19, 1524–1531 (2023). https://doi.org/10.1038/s41589-023-01402-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-023-01402-6