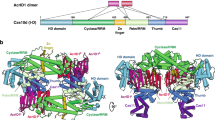
Abstract
CRISPR-Cas systems are prokaryotic antiviral systems, and phages use anti-CRISPR proteins (Acrs) to inactivate these systems. Here we present structural and functional analyses of AcrIF5, exploring its unique anti-CRISPR mechanism. AcrIF5 shows binding specificity only for the target DNA-bound form of the crRNA-guided surveillance (Csy) complex, but not the apo Csy complex from the type I-F CRISPR-Cas system. We solved the structure of the Csy–dsDNA–AcrIF5 complex, revealing that the conformational changes of the Csy complex caused by dsDNA binding dictate the binding specificity for the Csy–dsDNA complex by AcrIF5. Mechanistically, five AcrIF5 molecules bind one Csy–dsDNA complex, which destabilizes the helical bundle domain of Cas8f, thus preventing subsequent Cas2/3 recruitment. AcrIF5 exists in symbiosis with AcrIF3, which blocks Cas2/3 recruitment. This attack on the recruitment event stands in contrast to the conventional mechanisms of blocking binding of target DNA. Overall, our study reveals an unprecedented mechanism of CRISPR-Cas inhibition by AcrIF5.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The accession number for the coordinate and structure factor of AcrIF5 is PDB 7F45. The cryo-EM reconstruction of Csy–dsDNA–AcrIF5 has been deposited in the Electron Microscopy Data Bank under accession no. EMD-31265. Coordinates for the atomic model of Csy–dsDNA–AcrIF5 have been deposited in the Protein Data Bank under accession no. 7EQG. Structures of aPelota from A. pernix K1 (PDB 6JI2), Cas12i2 (PDB 6LTP), Tex protein from P. aeruginosa (PDB 3BZC) and Csy–dsDNA (PDB 6NE0) are referenced in the paper. Source data are provided with this paper.
References
Marraffini, L. A. CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61 (2015).
van der Oost, J., Westra, E. R., Jackson, R. N. & Wiedenheft, B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 12, 479–492 (2014).
Makarova, K. S. et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83 (2020).
Koonin, E. V., Makarova, K. S. & Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 37, 67–78 (2017).
Hille, F. et al. The biology of CRISPR-Cas: backward and forward. Cell 172, 1239–1259 (2018).
Borges, A. L., Davidson, A. R. & Bondy-Denomy, J. The discovery, mechanisms and evolutionary impact of anti-CRISPRs. Annu. Rev. Virol. 4, 37–59 (2017).
Bondy-Denomy, J. et al. A unified resource for tracking anti-CRISPR names. CRISPR J. 1, 304–305 (2018).
Pinilla-Redondo, R. et al. Discovery of multiple anti-CRISPRs highlights anti-defense gene clustering in mobile genetic elements. Nat. Commun. 11, 5652 (2020).
Marino, N. D. et al. Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science 362, 240–242 (2018).
Pawluk, A. et al. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat. Microbiol. 1, 16085 (2016).
Bondy-Denomy, J., Pawluk, A., Maxwell, K. L. & Davidson, A. R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2013).
Niu, Y. et al. A type I-F anti-CRISPR protein inhibits the CRISPR-Cas surveillance complex by ADP-ribosylation. Mol. Cell 80, 512–524 (2020).
Bondy-Denomy, J. et al. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526, 136–139 (2015).
Borges, A. L. et al. Bacteriophage cooperation suppresses CRISPR-Cas3 and Cas9 immunity. Cell 174, 917–925 (2018).
Rollins, M. F. et al. Structure reveals a mechanism of CRISPR-RNA-guided nuclease recruitment and anti-CRISPR viral mimicry. Mol. Cell 74, 132–142 (2019).
Chowdhury, S. et al. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell 169, 47–57 (2017).
Guo, T. W. et al. Cryo-EM structures reveal mechanism and inhibition of DNA targeting by a CRISPR-Cas surveillance complex. Cell 171, 414–426 (2017).
Zhang, K. et al. Inhibition mechanisms of AcrF9, AcrF8 and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM. Proc. Natl Acad. Sci. USA 117, 7176–7182 (2020).
Hirschi, M. et al. AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex. Nat. Commun. 11, 2730 (2020).
Gabel, C., Li, Z., Zhang, H. & Chang, L. Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14. Nucleic Acids Res. 49, 584–594 (2020).
Peng, R. et al. Alternate binding modes of anti-CRISPR viral suppressors AcrF1/2 to Csy surveillance complex revealed by cryo-EM structures. Cell Res. 27, 853–864 (2017).
Zhang, F. et al. CRISPRminer is a knowledge base for exploring CRISPR-Cas systems in microbe and phage interactions. Commun. Biol. 1, 180 (2018).
Pawluk, A. et al. Disabling a type I-E CRISPR-Cas nuclease with a bacteriophage-encoded anti-CRISPR protein. mBio 8, e01751-17 (2017).
Mejdani, M., Pawluk, A., Maxwell, K. L. & Davidson, A. R. Anti-CRISPR AcrIE2 binds the type I-E CRISPR-Cas complex but does not block DNA binding. J. Mol. Biol. 433, 166759 (2021).
Landsberger, M. et al. Anti-CRISPR phages cooperate to overcome CRISPR-Cas immunity. Cell 174, 908–916 (2018).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
The PyMOL Molecular Graphics System, Version 1.8 (Schrödinger, 2015).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Schindelin, J., Rueden, C. T., Hiner, M. C. & Eliceiri, K. W. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev. 82, 518–529 (2015).
Acknowledgements
We thank P. Wang at Southern University of Science and Technology for data collection. We acknowledge support with cryo-EM facilities from the Southern University of Science and Technology. We acknowledge the Protein Preparation and Characterization Core Facility of Tsinghua University Branch of China National Center for Protein Sciences Beijing and D. Li and Y. Li for providing the facility support. This work was supported by the National Natural Science Foundation of China (grants nos. 32171274 and 31822012), the National Key Research and Development Program of China (grant no. 2017YFA0506500), Beijing Nova program, the Fundamental Research Funds for the Central Universities (grant no. XK1802-8) to Y.F., the National Natural Science Foundation of China (grant no. 32000901), the National Key Research and Development Program of China (grants nos. 2019YFC1200500 and 2019YFC1200502) and the Beijing Natural Science Foundation (grant no. 5204038) to Y.Z., the State Key Program of National Natural Science of China (grant no. U1808202) and NSFC International (regional) Cooperation and Exchange Program (grant no. 31961143024) to Z.C.
Author information
Authors and Affiliations
Contributions
Y.F. conceived and supervised the project. Y.X., Z.G., P.Y., H.L. and H.W. purified the proteins, grew and optimized the crystals and performed the in vitro activity analysis and binding assays, supervised by Y.F. and Z.C. Y.F. collected the diffraction data and solved the crystal structure with the help of Y.Z. L.Z. collected the cryo-EM data and solved the cryo-EM structure, supervised by M.Y. Y.F. analyzed the data and wrote the manuscript, with assistance from all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Joseph Bondy-Denomy and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 AcrIF5 coelutes with the Csy-dsDNA complex but not the apo Csy.
Gel filtration profiles of the dsDNA and the mixes of Csy/dsDNA, Csy/AcrIF5 and Csy/dsDNA/AcrIF5. The UV absorbance at 280 nm is shown. The SDS-PAGE gels of the peak fractions of respective profile are shown on the right. The ‘*’ indicates contaminated proteins in the purified sample of the Csy complex. This experiment was performed 3 times with reproducible results.
Extended Data Fig. 2 Cryo-EM image processing for the Csy-dsDNA-AcrIF5 complex.
a, Representative processing pipeline includes a representative raw cryo-EM micrograph, 2D class averages, 3D classification and refinement of cryo-EM particles. b, Local resolution estimation and Euler angle distribution of the map of the Csy-dsDNA-AcrIF5 complex. c, Plot of the global FSC indicates an average resolution of 3.18 Å for the Csy-dsDNA-AcrIF5 complex.
Extended Data Fig. 3 Progressive displacement of Cas7f Subunits upon AcrIF5 binding to the Csy-dsDNA complex.
Comparison of each Cas7f subunit in Csy-dsDNA (PDB code: 6NE0, limon) and Csy-dsDNA-AcrIF5 (gray) complexes. The Cas7f subunits in the Csy-dsDNA complex become progressively displaced through mainly rigid-body movement as a result of AcrIF5 binding. Structures of the two complexes are aligned at Cas8f.
Extended Data Fig. 5 The crystal structure of AcrIF5.
a, Two AcrIF5 molecules are present in one asymmetric unit of the AcrIF5 crystal, colored in green and cyan. b, Structural superimposition of the two protomers of AcrIF5 in crystal. c, Structural superimposition of the two protomers of AcrIF5 in crystal (green and cyan) and the five AcrIF5 molecules in cryo-EM structure (colored as in Fig. 2a).
Extended Data Fig. 7 Structural mechanism of the binding specificity of AcrIF5 for the Csy-dsDNA complex.
a, Structural superimposition of the Cas7.2/7.3/7.4f subcomplex of the Csy (PDB code: 6B45) and Csy-dsDNA (PDB code: 6NE0), and the Cas7.2/7.3/7.4f-AcrIF5.3 subcomplex of the Csy-dsDNA-AcrIF5 (this study), aligned at the Cas7.2f subunit. b, Part of the structural superimposition in (a), in which only the Cas7.2/7.3/7.4f subcomplex of the Csy and AcrIF5.3 of the Cas7.2/7.3/7.4f-AcrIF5.3 subcomplex of the Csy-dsDNA-AcrIF5 are shown. Two clash regions are marked with circles. c, Part of the structural superimposition in (a), in which only the Cas7.2/7.3/7.4f subcomplex of the Csy-dsDNA and AcrIF5.3 of the Cas7.2/7.3/7.4f-AcrIF5.3 subcomplex of the Csy-dsDNA-AcrIF5 are shown. The corresponding regions of the two clash sites in (b) are marked with circles. d, On the basis of the structural superimposllition shown in (c), the Cas7.2f, 7.3f and 7.4f subunits in the structure of the Csy complex are aligned to their counterparts in the Csy-dsDNA structure. The same regions marked in (b) and (c) are also marked.
Extended Data Fig. 8 Structural superimposition of the Cas7f subunits of the Csy, Csy-dsDNA and Csy-dsDNA-AcrIF5 complexes.
a–f, Structural superimposition of the Cas7f subunits of the Csy (PDB code: 6B45, red), Csy-dsDNA (PDB code: 6NE0, limon) and Csy-dsDNA-AcrIF5 (this study, dark and light gray) complexes. The region of residues 232-243 of the web domain of Cas7.1f in the Csy-dsDNA-AcrIF5 complex is shown with cryo-EM density as blue mesh (a).
Extended Data Fig. 9 AcrIF5 coexists with AcrIF3 in the same operon.
Shown is a schematic representation of the genomic context of acrIF5 genes. Genes shown in the same color represent the same family. Genes encoding an HTH-containing protein, a phage virion morphogenesis protein and an SOS response-associated peptidase are colored green, gray and light pink, respectively. Arrows representing genes are not shown to scale.
Extended Data Fig. 10 MST binding assays of AcrIF5 and Cas2/3 to the Csy-dsDNA complex.
MST assay of the binding between the purified Csy-dsDNA complex and AcrIF5 (a) or Cas2/3 (b). The binding curves are displayed. Shown are mean values ± s.d.; n=3 biologically independent experiments. Individual data points are also shown.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5 and Tables 1 and 2.
Source data
Source Data Fig. 1
Uncropped gel.
Rights and permissions
About this article
Cite this article
Xie, Y., Zhang, L., Gao, Z. et al. AcrIF5 specifically targets DNA-bound CRISPR-Cas surveillance complex for inhibition. Nat Chem Biol 18, 670–677 (2022). https://doi.org/10.1038/s41589-022-00995-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-00995-8
This article is cited by
-
Insights into the inhibition of protospacer integration via direct interaction between Cas2 and AcrVA5
Nature Communications (2024)