Abstract
Cancer cells have long been recognized to exhibit unique bioenergetic requirements. The apoptolidin family of glycomacrolides are distinguished by their selective cytotoxicity towards oncogene-transformed cells, yet their molecular mechanism remains uncertain. We used photoaffinity analogs of the apoptolidins to identify the F1 subcomplex of mitochondrial ATP synthase as the target of apoptolidin A. Cryogenic electron microscopy (cryo-EM) of apoptolidin and ammocidin–ATP synthase complexes revealed a novel shared mode of inhibition that was confirmed by deep mutational scanning of the binding interface to reveal resistance mutations which were confirmed using CRISPR–Cas9. Ammocidin A was found to suppress leukemia progression in vivo at doses that were tolerated with minimal toxicity. The combination of cellular, structural, mutagenesis, and in vivo evidence defines the mechanism of action of apoptolidin family glycomacrolides and establishes a path to address oxidative phosphorylation-dependent cancers.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The atomic coordinates have been deposited in the Protein Data Bank (PDB) with the accession codes 7MD2 and 7MD3. The EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) with the accession codes 23763, 23764 and 23765. The sequencing data and variant counts for the deep mutational scanning experiments have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE171362. The data generated in this study are contained in the published article including its Supplementary information. Source data are provided with this paper.
Code availability
Code used for data analysis generation of figures is avalible at github.com/bjreisman/2021_NCB_apoptolidin.
Material availability
Materials and compounds are available from the corresponding author upon request.
References
Akimov, Y. & Aittokallio, T. Re-defining synthetic lethality by phenotypic profiling for precision oncology. Cell Chem. Biol. 28, 246–256 (2021).
Pavlova, N. N. & Thompson, C. B. The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
DeBerardinis, R. J. & Chandel, N. S. We need to talk about the Warburg effect. Nat. Metab. 2, 127–129 (2020).
Xu, Y., Xue, D., Bankhead, A. 3rd & Neamati, N. Why all the fuss about oxidative phosphorylation (OXPHOS)? J. Med. Chem. 63, 14276–14307 (2020).
Martinez-Reyes, I. et al. Mitochondrial ubiquinol oxidation is necessary for tumour growth. Nature 585, 288–292 (2020).
Jones, C. L., Inguva, A. & Jordan, C. T. Targeting energy metabolism in cancer stem cells: progress and challenges in leukemia and solid tumors. Cell Stem Cell 28, 378–393 (2021).
Carter, J. L. et al. Targeting mitochondrial respiration for the treatment of acute myeloid leukemia. Biochem. Pharmacol. 182, 114253 (2020).
Lagadinou, E. D. et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 12, 329–341 (2013).
Pei, S. et al. Monocytic subclones confer resistance to venetoclax-based therapy in patients with acute myeloid leukemia. Cancer Discov. 10, 536–551 (2020).
Sighel, D. et al. Inhibition of mitochondrial translation suppresses glioblastoma stem cell growth. Cell Rep. 35, 109024 (2021).
Wang, F. et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 340, 622–626 (2013).
Faubert, B., Solmonson, A. & Deberardinis, R. J. Metabolic reprogramming and cancer progression. Science 368, eaaw5473 (2020).
Kim, J. W., Adachi, H., Shin-ya, K., Hayakawa, Y. & Seto, H. Apoptolidin, a new apoptosis inducer in transformed cells from Nocardiopsis sp. J. Antibiot. (Tokyo) 50, 628–630 (1997).
Murakami, R. et al. Ammocidin, a new apoptosis inducer in Ras-dependent cells from Saccharothrix sp. I. Production, isolation and biological activity. J. Antibiot. (Tokyo) 54, 710–713 (2001).
Salomon, A. R., Voehringer, D. W., Herzenberg, L. A. & Khosla, C. Apoptolidin, a selective cytotoxic agent, is an inhibitor of F0F1-ATPase. Chem. Biol. 8, 71–80 (2001).
Wender, P. A. et al. Correlation of F0F1-ATPase inhibition and antiproliferative activity of apoptolidin analogues. Org. Lett. 8, 589–592 (2006).
Serrill, J. D. et al. Apoptolidins A and C activate AMPK in metabolically sensitive cell types and are mechanistically distinct from oligomycin A. Biochem. Pharmacol. 93, 251–265 (2015).
DeGuire, S. M. et al. Fluorescent probes of the apoptolidins and their utility in cellular localization studies. Angew. Chem. Int. Ed. 54, 961–964 (2015).
Mackinnon, A. L. & Taunton, J. Target identification by diazirine photo-cross-linking and click chemistry. Curr. Protoc. Chem. Biol. 1, 55–73 (2009).
Du, Y. et al. Biosynthesis of the apoptolidins in Nocardiopsis sp. FU 40. Tetrahedron 67, 6568–6575 (2011).
Salomon, A. R., Voehringer, D. W., Herzenberg, L. A. & Khosla, C. Understanding and exploiting the mechanistic basis for selectivity of polyketide inhibitors of F0F1-ATPase. Proc. Natl Acad. Sci. USA 97, 14766–14771 (2000).
Wender, P. A., Jankowski, O. D., Tabet, E. A. & Seto, H. Toward a structure–activity relationship for apoptolidin: selective functionalization of the hydroxyl group array. Org. Lett. 5, 487–490 (2003).
Abrahams, J. P., Leslie, A. G., Lutter, R. & Walker, J. E. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 (1994).
Guo, H. & Rubinstein, J. L. Cryo-EM of ATP synthases. Curr. Opin. Struct. Biol. 52, 71–79 (2018).
West, A. V. et al. Labeling preferences of diazirines with protein biomolecules. J. Am. Chem. Soc. 143, 6691–6700 (2021).
Salomon, A. R., Zhang, Y., Seto, H. & Khosla, C. Structure–activity relationships within a family of selectively cytotoxic macrolide natural products. Org. Lett. 3, 57–59 (2001).
Gledhill, J. R. & Walker, J. E. Inhibitors of the catalytic domain of mitochondrial ATP synthase. Biochem. Soc. Trans. 34, 989–992 (2006).
Symersky, J., Osowski, D., Walters, D. E. & Mueller, D. M. Oligomycin frames a common drug-binding site in the ATP synthase. Proc. Natl Acad. Sci. USA 109, 13961–13965 (2012).
Tantama, M., Martinez-Francois, J. R., Mongeon, R. & Yellen, G. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat. Commun. 4, 2550 (2013).
Gledhill, J. R., Montgomery, M. G., Leslie, A. G. & Walker, J. E. How the regulatory protein, IF1, inhibits F1-ATPase from bovine mitochondria. Proc. Natl Acad. Sci. USA 104, 15671–15676 (2007).
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Matreyek, K. A., Stephany, J. J., Chiasson, M. A., Hasle, N. & Fowler, D. M. An improved platform for functional assessment of large protein libraries in mammalian cells. Nucleic Acids Res. 48, e1 (2020).
Wrenbeck, E. E. et al. Plasmid-based one-pot saturation mutagenesis. Nat. Methods 13, 928–930 (2016).
Smith, R. M. Biological and chemical studies on a new antibiotic, oligomycin, University of Wisconsin–Madison, (1953).
Pennington, J. D., Williams, H. J., Salomon, A. R. & Sulikowski, G. A. Toward a stable apoptolidin derivative: identification of isoapoptolidin and selective deglycosylation of apoptolidin. Org. Lett. 4, 3823–3825 (2002).
Ramsey, H. E. et al. A novel MCL1 inhibitor combined with venetoclax rescues venetoclax-resistant acute myelogenous leukemia. Cancer Discov. 8, 1566–1581 (2018).
Baccelli, I. et al. Mubritinib targets the electron transport chain complex I and reveals the landscape of OXPHOS dependency in acute myeloid leukemia. Cancer Cell 36, 84–99.e8 (2019).
Jones, C. L. et al. Inhibition of amino acid metabolism selectively targets human leukemia stem cells. Cancer Cell 34, 724–740.e4 (2018).
Pollyea, D. A. et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat. Med. 24, 1859–1866 (2018).
Sriskanthadevan, S. et al. AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood 125, 2120–2130 (2015).
Farge, T. et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 7, 716–735 (2017).
Ashton, T. M., McKenna, W. G., Kunz-Schughart, L. A. & Higgins, G. S. Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res. 24, 2482–2490 (2018).
Molina, J. R. et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med. 24, 1036–1046 (2018).
Sharon, D. et al. Inhibition of mitochondrial translation overcomes venetoclax resistance in AML through activation of the integrated stress response. Sci. Transl. Med. 11, eaax2863 (2019).
Guieze, R. et al. Mitochondrial reprogramming underlies resistance to BCL-2 inhibition in lymphoid malignancies. Cancer Cell 36, 369–384 e13 (2019).
Gorelick, A. N. et al. Respiratory complex and tissue lineage drive recurrent mutations in tumour mtDNA. Nat. Metab. 3, 558–570 (2021).
Arrowsmith, C. H. et al. The promise and peril of chemical probes. Nat. Chem. Biol. 11, 536–541 (2015).
Moulton, M. J. & Letsou, A. Modeling congenital disease and inborn errors of development in Drosophila melanogaster. Dis. Model. Mech. 9, 253–269 (2016).
Mueller, D. M. et al. Ni-chelate-affinity purification and crystallization of the yeast mitochondrial F1-ATPase. Protein Expr. Purif. 37, 479–485 (2004).
Rubinstein, J. L., Dickson, V. K., Runswick, M. J. & Walker, J. E. ATP synthase from Saccharomyces cerevisiae: location of subunit h in the peripheral stalk region. J. Mol. Biol. 345, 513–520 (2005).
Marr, C. R., Benlekbir, S. & Rubinstein, J. L. Fabrication of carbon films with approximately 500 nm holes for cryo-EM with a direct detector device. J. Struct. Biol. 185, 42–47 (2014).
Tivol, W. F., Briegel, A. & Jensen, G. J. An improved cryogen for plunge freezing. Microsc. Microanal. 14, 375–379 (2008).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Medina-Cucurella, A. V. et al. User-defined single pot mutagenesis using unamplified oligo pools. Protein Eng. Des. Sel. 32, 41–45 (2019).
Boyce, J. H., Reisman, B. J., Bachmann, B. O. & Porco, J. A. Jr. Synthesis and multiplexed activity profiling of synthetic acylphloroglucinol scaffolds. Angew. Chem. Int. Ed. 60, 1263–1272 (2021).
Baldwin, C. Biological and chemical properties of aurovertin, a metabolic product of Calcarisporium abuscula. Lloydia 27, 88–95 (1964).
Krasnoff, S. B. & Gupta, S. Identification and directed biosynthesis of efrapeptins in the fungus Tolypocladium geodes Gams (Deuteromycotina: Hyphomycetes). J. Chem. Ecol. 17, 1953–1962 (1991).
Acknowledgements
We are grateful to K. Matreyek of Case Western Reserve University for providing the Bxb1 landing pad cells and plasmids for the mutagenesis experiments, and A. Muhammad and A. Glazer for assistance applying the landing pad system. We are grateful to D. Mueller of Rosalind Franklin University for providing the USY006 yeast strain and protocols for handling isolated mitochondria, as well as P. Sharma for his assistance isolating F1 ATPase for enzymatic assays. We thank K. Brown and H.-Y. Kim for their helpful advice regarding synthesis and affinity enrichment, as well as A. Gorska for her assistance with immunohistochemistry experiments. The ammocidin producing strain AJ9571 was provided by Ajinomoto Co., Inc., Tokyo, Japan under a material transfer agreement. This work was supported by research grants from the National Institutes of Health for research (R01 GM092218 to B.O.B., R01 CA226833 to B.J.R. and B.O.B., R35 GM133552 to L.P., K23 HL138291 to P.B.F.), and training support (T32 GM065086 (B.J.R., M.T.W.), T32 GM007347 (B.J.R., B.I.R.), F30 CA236131 (B.J.R.), F30 CA247202 (B.I.R.). This work was also supported by a Canadian Institutes of Health grant PJT162186 (J.L.R.). J.L.R. was supported by the Canada Research Chairs program and H.G. by an International Student Ontario Graduate Scholarship. M.T.W was supported by the National Science Foundation Graduate Research Fellowship Program. We are thankful for the resources provided by the following core facilities: Vanderbilt Flow Cytometry Shared Resource (supported by the Vanderbilt Ingram Cancer Center (P30 CA68485)), Vanderbilt VANTAGE (VANTAGE is supported in part by CTSA Grant (5UL1 RR024975-03)) genomics core, the Vanderbilt Cell Imaging Shared Resource (CISR is supported by NIH grant DK020593), Vanderbilt Small Molecule NMR Facility core (supported in part by NIH grant S10 RR019022), and Center for Innovation Technology at Vanderbilt University. Cryo-EM data was collected at the Toronto High-Resolution High-Throughput cryo-EM facility, supported by the Canada Foundation for Innovation and Ontario Research Fund.
Author information
Authors and Affiliations
Contributions
B.J.R., G.A.S, L.P. and B.O.B. conceived of the project. B.J.R., H.G., H.E.R., M.T.W., B.I.R. and P.B.F. designed the experiments. B.J.R. isolated compounds, sythesized and validated probes, performed affinity enrichments and gel-based profiling, and carried out mutational scanning and resistance experiements. H.G. prepared cryo-EM grids, collected and analyzed sturctural data. H.E.R. performed xenograft experiments. M.T.W. carried out and analyzed proteomics experiments. B.I.R. generated stable cells lines and conducted toxicity studies. All authors contributed to the data analysis. B.J.R. and H.G. wrote the manuscript. P.B.F., W.K.R., L.P., M.R.S., J.L.R. and B.O.B. supervised the project.
Corresponding author
Ethics declarations
Competing interests
M.R.S. receives research funding from Astex, Incyte, Takeda and TG Therapeutics; has equity with Karyopharm; serves as an advisor or consultant to AbbVie, Astex, BMS, Celgene, Incyte, Karyopharm, Ryvu, Sierra Oncology, Takeda and TG Therapeutics. P.B.F. currently receives research funding from Incyte, and has received research funding from Astex and Forma Therapeutics in the past. Within the past 2 years, W.K.R. has received unrelated clinical research support Bristol-Meyers Squib, Merck, Pfizer, Peloton, Calithera and Incyte.
Additional information
Peer review information Nature Chemical Biology thanks Alexey Amunts, Markus Schirle and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Extended validation of glycomacrolide photoaffinity probes for target identification.
a, Analysis of pS6 phosphorylation using FCB barcoded flow cytometry of MV-4-11 cells treated for 16 h with glycomacrolides or probes; b, Concentrations response curves in MV-4-11 cells treated for 72 h with Apop H/Apop H-PA and c, Ammo A/Ammo A-PA; d, Gel-based profiling of Apop H-PA or e, Ammo A-PA adducts in MV-4-11 cells; f, Gel-based profiling of Apop A-PA targets in BaF3 cells transformed with HRas-G12V; g Immunoblot showing specific enrichment of ATP5B in H292 cells treated with Apop A-PA; h; Gel-based analysis of diazirine and UV light dependent labeling of Apop A-PA at 1 μM.
Extended Data Fig. 2 Identification of ATP5B as the target of apoptolidin A using comparative proteomics.
a, Gel-based profiling of Apop A-PA targets in MV-4-11 with ± 1 μM Apop A-PA, ± 20 μM Apop A after affinity enrichment using streptavidin resin prior to digestion and TMT labeling, imaged by rhodamine fluorescence and after silver staining; b, schematic of the TMT multiplexing strategy for to combine vehicle, probe, and competition, conditions from two separate biological replicates; c, Volcano plot of proteins significantly enriched in ‘Probe’ condition compared to ‘Vehicle’ analyzed by TMT-multiplexed MudPIT proteomics; d, Volcano plot of proteins significantly enriched in ‘Competition’ condition compared to ‘Vehicle’ analyzed by TMT-multiplexed MuDPIT proteomics; e, Coomassie stained gel-based profiling of Apop A-PA targets in MV-4-11 cells, the 50 kDa band from probe and competition conditions were cut and subjected to in-gel digestion (see Supplementary Table 1); f, Immunoblot showing pulldown of FLAG tagged ATP5B expressed in HEK-293 landing pad cells.
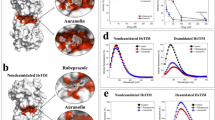
Extended Data Fig. 3 Analysis of apoptolidin A binding mode via photoaffinity labeling and enzymatic assays.
a, Concentration response-curves showing inhibition of ATPase activity in purified yeast FOF1 ATP synthase, measured using PK/LDH coupled assay; b, Gel-based analysis of Apop A competition against Apop A-PA at a fixed concentration; c, Gel-based analysis of Apop A-PA in the presence of known ATP synthase inhibitors Aurovertin B (Auro B), Efrapeptin F (Efra F), Citreoviridin (Citr), and Oligomycin A (Oligm); d, Analysis of ADP concentration dependence on inhibition of ATP synthesis by Apop A in isolated mouse liver mitochondria using HK/G6PDH coupled assay; e, Analysis of ATP concentration dependence on inhibition of ATP hydrolysis by Apop A and Ammo A in isolated yeast F1 ATPase using a PK/LDH coupled assay; f, Gel-based profiling of Apop A-PA in MV-4-11 cells treated with or without uncoupling agents (1 μM); g, confirmation of ATPIF1 KO in two independent K562 clones after knockout using PX458 with two independent ATP5IF1 targeting gRNAs; h, Gel-based profiling of membrane potential dependent adduction of Apop A-PA in WT or IF1 KO K562 cells; i, comparison of ammocidin bound (left) to IF1 bound F1 ATPase.
Extended Data Fig. 4 Workflow for cryo-EM image analysis and validation for ammocidin-bound and apoptolidin-bound yeast ATP synthase.
Example micrographs (a, c), 2D class average images (b, d), and workflow for obtaining maps of the F1 regions (e, f) for ammocidin-bound, and apoptolidin-bound ATP synthases, respectively. Corrected Fourier shell correlation curves after gold-standard refinement g, local resolution maps h, and orientation distribution plots i, are shown for the maps of the F1 regions of the ammocidin-bound, apoptolidin-bound, and inhibitor-free data sets. Alternative conformations of ammocidin (j) and apoptolidin (k) atomic models.
Extended Data Fig. 5 ATP5B-L394F mutation has opposing effects on apoptolidin and ammocidin sensitivity.
a, Multiple sequence alignment of ATP synthase β subunit C-terminal domain and CRISPR–Cas9 HDR editing strategy for human exon 9 and using plasmid (PX458) encoding sgRNA and Cas9 and ssODN repair template, PAM sites labeled in yellow, cut locations noted with arrowhead, edits noted in red, restriction sites noted; b, Restriction digest of ATP5F1B Exon 9/10 amplicons from parental K562s or PX458 + ssODN treated K562s treated with Ammo A (150 nM x 5 d) showing loss of wildtype allele and retention of edited alleles with ammocidin treatment; c, Confirmation of introduction of ATP5B-L394F mutation and KO of wildtype ATP5B in K562s and MV-4-11 cells by Sanger sequencing; d, ATP5B-L394F sensitizes cells to Apop A and provides some resistance against ammocidin in K562 and e, MV-4-11 cells as measured by MTT assay at 72 h; f, Modeling of the L394 (yeast L397) mutation on the ammocidin (top) and apoptolidin (bottom) structures showing proximity to the hemiketal.
Extended Data Fig. 6 Deep mutational scanning of the ammocidin binding site on ATP Synthase.
a, Selection of residues for mutagenesis on α (Yeast ATP1, Human ATP5A – red), β (Yeast ATP2, Human ATP5B – yellow), and γ (Yeast ATP3, Human ATP5C – blue) subunits within 4.5 Å of ammocidin or apoptolidin; b, c, Validation of ATP synthase variant library after nicking mutagenesis showing the frequency of non-wildtype variants at each position, with residue 1 corresponding to the first residue of the mature peptide; d, Confirmation of proper mitochondrial localization of the landing pad expressed ATP5A by immunofluorescence at 63X in HEK-293T landing pad cells; e, Assessment of reproducibility between biological replicates showing consistent enrichment of resistance mutations, variants which were not observed were set to −2.5; f, Comparison of variant frequencies observed after selection for successful integration compared to the original library, variants which were observed in the original library but not detected after integration were set to -Inf. Variants which exhibit negative enrichment or complete loss are thought to exhibit decreased fitness relative to the WT alleles; g, Comparison of variants enriched by apoptolidin and ammocidin at medium dose (100 nM / 30 nM) or h, high (1 μM / 300 nM) demonstrating that apoptolidin and ammocidin select for similar resistance mutations.
Extended Data Fig. 7 Apoptolidin and ammocidin select for similar resistance mutations.
Tile maps of amino acid enrichment relative to the parental library at each position at each dose of ammocidin or apoptolidin; a, Vehicle treated cells were allowed to grow for one additional passage after selection, while remaining wells were passaged with Apoptolidin A at concentrations of b, 100 nM or c, 1 μM or Ammocidin A at concentrations of d, 3 nM, e, 30 nM, f, 300 nM. White squares indicate variants which were not detected (<100 counts) by deep sequencing, black circles represent WT alleles.
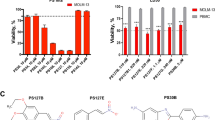
Extended Data Fig. 8 Validation of ATP synthase resistance mutations using CRISPR–Cas9 genome editing.
a, Validation of resistance mutations using transgenic mutant ATP synthase in HEK-293-LP cells at 72 h by SRB assay for mutations in the alpha, b, beta, and c, gamma subunits; d, CRISPR–Cas9 HDR editing strategy for ATP5B exon 9 and e, ATP5C exon 4 using plasmid (PX458) encoding sgRNA and Cas9 and ssODN repair template, PAM sites labeled in yellow, cut locations noted with arrowhead, edits noted in red; f, Confirmation of introduction of ATP5B-I390R in K562 cells, g, ATP5C-L77R in MV-4-11 cells, or h, ATP5C-L77R in K562 cells by sanger sequencing of single-cell clones via PCR amplicons and/or PCR cloning to resolve heterozygous edits; i, Confirmation of unchanged expression of ATP synthase genes in CRISPR–Cas9 edited clones by immunoblot; j, Secondary validation of ATP5C-L77R mutant in CRISPR edited MV-4-11 cell line using MTT-assay at 48 h; k, Gel-based profiling of Apop A-PA targets in K562s Parental or CRISPR–Cas9 edited cell lines showing loss of binding to ATP5B visualized using TAMRA-azide-desthiobiotin.
Extended Data Fig. 9 Evaluation of ammocidin dosing, pharmacokinetics and toxicity in NSGS mice and efficacy in NSGS MV-4-11 xenograft.
a, Preliminary (non-GLP) evaluation of PK profile of Ammo A in NSGS mice with a single dose of Ammo A (technical duplicate); b, Gross histology of H&E sections from organs of NSGS mice 2 weeks after treatment with Ammo A (0.1 mg/kg IP QD for 5 on, 2 off) revealing/reveal no tissue damage related toxicity; c, Engrafted mice (n = 4 per group) were treated from day 7 to day 18 (pink brackets) and therapy was then held. Mouse survival was measured by Kaplan-Meier analysis (P value calculated using two-sided log-rank test.).
Extended Data Fig. 10 Gating schemes for flow cytometry data.
Gating schemes for a. Perceval-HR reporter MV-4-11s; b, multiplexed activity profiling in MV-4-11 cells; c, ATPIF1 KO K562 cells; and d, murine xenograft chimerism experiments.
Supplementary information
Supplementary Information
Supplementary Cryo-EM modeling data, primer sequences, chemical synthesis methods and NMR spectra.
Supplementary Table 1
Proteomics data from affinity enrichments of apoptolidin A-PA adducts in MV-4-11 cells.
Supplementary Video 1
Structure of ammocidin-ATP synthase.
Source data
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Reisman, B.J., Guo, H., Ramsey, H.E. et al. Apoptolidin family glycomacrolides target leukemia through inhibition of ATP synthase. Nat Chem Biol 18, 360–367 (2022). https://doi.org/10.1038/s41589-021-00900-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00900-9
This article is cited by
-
Bioinformatic and literature assessment of toxicity and allergenicity of a CRISPR-Cas9 engineered gene drive to control Anopheles gambiae the mosquito vector of human malaria
Malaria Journal (2023)
-
Pan-tissue mitochondrial phenotyping reveals lower OXPHOS expression and function across cancer types
Scientific Reports (2023)
-
“Proteotranscriptomic analysis of advanced colorectal cancer patient derived organoids for drug sensitivity prediction”
Journal of Experimental & Clinical Cancer Research (2023)
-
A wrench in the motor
Nature Chemical Biology (2022)