Abstract
The neuropeptide substance P (SP) is important in pain and inflammation. SP activates the neurokinin-1 receptor (NK1R) to signal via Gq and Gs proteins. Neurokinin A also activates NK1R, but leads to selective Gq signaling. How two stimuli yield distinct G protein signaling at the same G protein-coupled receptor remains unclear. We determined cryogenic-electron microscopy structures of active NK1R bound to SP or the Gq-biased peptide SP6–11. Peptide interactions deep within NK1R are critical for receptor activation. Conversely, interactions between SP and NK1R extracellular loops are required for potent Gs signaling but not Gq signaling. Molecular dynamics simulations showed that these superficial contacts restrict SP flexibility. SP6–11, which lacks these interactions, is dynamic while bound to NK1R. Structural dynamics of NK1R agonists therefore depend on interactions with the receptor extracellular loops and regulate G protein signaling selectivity. Similar interactions between other neuropeptides and their cognate receptors may tune intracellular signaling.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its Supplementary Information. Coordinates for SP–NK1R–miniGs/q70, SP–NK1R–miniGs399 and the SP6–11–NK1R–miniGs/q70 complex have been deposited in the PDB under accession codes 7RMG, 7RMH and 7RMI, respectively. Unsharpened and sharpened EM density maps and half maps for SP–NK1R–miniGs/q70, SP–NK1R–miniGs399 and the SP6–11–NK1R–miniGs/q70 complex have been deposited in the Electron Microscopy Data Bank under accession codes 24569, 24570 and 24572, respectively. Final particle stacks for SP–NK1R–miniGs/q70, SP–NK1R–miniGs399, and the SP6–11–NK1R–miniGs/q70 complex reconstruction have been uploaded to the Electron Microscopy Public Image Archive under the accession code EMD-EMPIAR 10786. Simulation trajectories for molecular dynamics simulations have been deposited on Zenodo, and are available at https://doi.org/10.5281/zenodo.5113874Source data are provided with this paper.
References
Steinhoff, M. S., von Mentzer, B., Geppetti, P., Pothoulakis, C. & Bunnett, N. W. Tachykinins and their receptors: contributions to physiological control and the mechanisms of disease. Physiol. Rev. 94, 265–301 (2014).
Hargreaves, R. et al. Development of aprepitant, the first neurokinin-1 receptor antagonist for the prevention of chemotherapy-induced nausea and vomiting. Ann. NY Acad. Sci. 1222, 40–48 (2011).
Maggi, C. A. The mammalian tachykinin receptors. Gen. Pharmacol. 26, 911–944 (1995).
Schwyzer, R. ACTH: a short introductory review. Ann. NY Acad. Sci. 297, 3–26 (1977).
Hökfelt, T., Pernow, B. & Wahren, J. Substance P: a pioneer amongst neuropeptides. J. Intern. Med. 249, 27–40 (2001).
Mazzone, S. B. & Geraghty, D. P. Respiratory actions of tachykinins in the nucleus of the solitary tract: characterization of receptors using selective agonists and antagonists. Br. J. Pharmacol. 129, 1121–1131 (2000).
Trafton, J. A., Abbadie, C. & Basbaum, A. I. Differential contribution of substance P and neurokinin A to spinal cord neurokinin-1 receptor signaling in the rat. J. Neurosci. 21, 3656–3664 (2001).
Sagan, S., Chassaing, G., Pradier, L. & Lavielle, S. Tachykinin peptides affect differently the second messenger pathways after binding to CHO expressed human NK1 receptors. J. Pharmacol. Exp. Ther. 276, 1039–1048 (1996).
Riitano, D., Werge, T. M. & Costa, T. A mutation changes ligand selectivity and transmembrane signaling preference of the neurokinin-1 receptor. J. Biol. Chem. 272, 7646–7655 (1997).
Evans, T. W., Dixon, C. M., Clarke, B., Conradson, T. B. & Barnes, P. J. Comparison of neurokinin A and substance P on cardiovascular and airway function in man. Br. J. Clin. Pharmacol. 25, 273–275 (1988).
Hastrup, H. & Schwartz, T. W. Septide and neurokinin A are high-affinity ligands on the NK-1 receptor: evidence from homologous versus heterologous binding analysis. FEBS Lett. 399, 264–266 (1996).
Ciucci, A., Palma, C., Manzini, S. & Werge, T. M. Point mutation increases a form of the NK receptor with high affinity for neurokinin A and B and septide. Br. J. Pharmacol. 125, 393–401 (1998).
Maggi, C. A. & Schwartz, T. W. The dual nature of the tachykinin NK1 receptor. Trends Pharmacol. Sci. 18, 351–355 (1997).
Nehmé, R. et al. Mini-G proteins: novel tools for studying GPCRs in their active conformation. PLoS ONE 12, e0175642 (2017).
Rasmussen, S. G. F. et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011).
Ballesteros, J. A. & Weinstein, H. in Receptor Molecular Biology (ed. Sealfon, S. C.) Ch. 19, 366–428 (Academic Press, 1995).
Huang, W. et al. Structural insights into µ-opioid receptor activation. Nature 524, 315–321 (2015).
Manglik, A. et al. Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. Nature 485, 321–326 (2012).
Kim, K. et al. Structure of a hallucinogen-activated Gq-coupled 5-HT2A serotonin receptor. Cell 182, 1574–1588.e19 (2020).
Maeda, S., Qu, Q., Robertson, M. J., Skiniotis, G. & Kobilka, B. K. Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 364, 552–557 (2019).
García-Nafría, J., Lee, Y., Bai, X., Carpenter, B. & Tate, C. G. Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. eLife 7, e35946 (2018).
Kato, H. E. et al. Conformational transitions of a neurotensin receptor 1-Gi1 complex. Nature 572, 80–85 (2019).
Egloff, P. et al. Structure of signaling-competent neurotensin receptor 1 obtained by directed evolution in Escherichia coli. Proc. Natl Acad. Sci. USA 111, E655–E662 (2014).
Schöppe, J. et al. Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists. Nat. Commun. 10, 17 (2019).
Valentin-Hansen, L., Frimurer, T. M., Mokrosinski, J., Holliday, N. D. & Schwartz, T. W. Biased Gs versus Gq proteins and β-arrestin signaling in the NK1 receptor determined by interactions in the water hydrogen bond network. J. Biol. Chem. 290, 24495–24508 (2015).
Koehl, A. et al. Structure of the µ-opioid receptor-Gi protein complex. Nature 558, 547–552 (2018).
Hong, C. et al. Structures of active-state orexin receptor 2 rationalize peptide and small-molecule agonist recognition and receptor activation. Nat. Commun. 12, 815 (2021).
Yin, J. et al. Crystal structure of the human NK1 tachykinin receptor. Proc. Natl Acad. Sci. USA 115, 13264–13269 (2018).
Fong, T. M. et al. Mutational analysis of neurokinin receptor function. Can. J. Physiol. Pharmacol. 73, 860–865 (1995).
Lane, J. R., May, L. T., Parton, R. G., Sexton, P. M. & Christopoulos, A. A kinetic view of GPCR allostery and biased agonism. Nat. Chem. Biol. 13, 929–937 (2017).
Holst, B., Hastrup, H., Raffetseder, U., Martini, L. & Schwartz, T. W. Two active molecular phenotypes of the tachykinin NK1 receptor revealed by G-protein fusions and mutagenesis. J. Biol. Chem. 276, 19793–19799 (2001).
Wan, Q. et al. Mini G protein probes for active G protein–coupled receptors (GPCRs) in live cells. J. Biol. Chem. 293, 7466–7473 (2018).
Nederpelt, I., Bleeker, D., Tuijt, B., IJzerman, A. P. & Heitman, L. H. Kinetic binding and activation profiles of endogenous tachykinins targeting the NK1 receptor. Biochem. Pharmacol. 118, 88–95 (2016).
Suomivuori, C.-M. et al. Molecular mechanism of biased signaling in a prototypical G protein-coupled receptor. Science 367, 881–887 (2020).
Wheatley, M. et al. Lifting the lid on GPCRs: the role of extracellular loops. Br. J. Pharmacol. 165, 1688–1703 (2012).
Thompson, G. L. et al. Biased agonism of endogenous opioid peptides at the μ-Opioid receptor. Mol. Pharmacol. 88, 335–346 (2015).
Liao, Y. et al. Human neuropeptide S receptor is activated via a Gαq protein-biased signaling cascade by a human neuropeptide S analog lacking the C-terminal 10 residues. J. Biol. Chem. 291, 7505–7516 (2016).
Kaiser, A., Wanka, L., Ziffert, I. & Beck-Sickinger, A. G. Biased agonists at the human Y1 receptor lead to prolonged membrane residency and extended receptor G protein interaction. Cell. Mol. Life Sci. 77, 4675–4691 (2020).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Asarnow, D., Palovcak, E. & Cheng, Y. asarnow/pyem: UCSF pyem v.0.5 https://doi.org/10.5281/zenodo.3576630 (Zenodo, 2019).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D. Struct. Biol. 75, 861–877 (2019).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Ghanouni, P. et al. The effect of pH on β2 adrenoceptor function: evidence for protonation-dependent activation. J. Biol. Chem. 275, 3121–3127 (2000).
Ranganathan, A., Dror, R. O. & Carlsson, J. Insights into the role of Asp79(2.50) in β2 adrenergic receptor activation from molecular dynamics simulations. Biochemistry 53, 7283–7296 (2014).
Zhang, L. & Hermans, J. Hydrophilicity of cavities in proteins. Proteins: Struct. Funct. Genet. 24, 433–438 (1996).
Lomize, M. A., Lomize, A. L., Pogozheva, I. D. & Mosberg, H. I. OPM: orientations of proteins in membranes database. Bioinformatics 22, 623–625 (2006).
Betz, R. M. Dabble https://doi.org/10.5281/zenodo.836914 (Zenodo, 2017).
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 114, 7830–7843 (2010).
Case, D. A. et al. AMBER 2018 (Univ. California San Francisco, 2018).
Salomon-Ferrer, R. et al. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 8, 1542–1555 (2012).
Hopkins, C. W., Le Grand, S., Walker, R. C. & Roitberg, A. E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 11, 1864–1874 (2015).
Roe, D. R. & Cheatham, T. E. 3rd PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 27–28 (1996).
Michaud-Agrawal, N., Denning, E. J., Woolf, T. B. & Beckstein, O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32, 2319–2327 (2011).
Acknowledgements
We thank M. Yeasmin for assisting with IP1 accumulation assays. This work was supported by National Institutes of Health (NIH) grant nos. 1R35GM140847 (Y.C.), DP5OD023048 (A.M.), R01GM127359 (R.O.D.) and a National Health and Medical Research Council (NHMRC) Project Grant APP1138448 (D.M.T.), NHMRC Investigator grant no. APP1196951 (D.M.T), Australian Research Council DECRA grant no. DE170100152 (D.M.T.) and Australian Research Council Centre of Excellence in Convergent Bio-Nano Science and Technology (N.A.V.). This material is based on work supported by the National Science Foundation Graduate Research Fellowship Program (J.A.H.) under grant no. 2034836. Any opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. C.-M.S. was funded by the Human Frontier Science Program (grant no. LT000916/2018-L). Cryo-EM equipment at UCSF is partially supported by NIH grant nos. S10OD020054 and S10OD021741. Some of this work was performed at the Stanford-SLAC Cryo-EM Center (S2C2), which is supported by the NIH Common Fund Transformative High-Resolution Cryo-Electron Microscopy program (grant no. U24 GM129541). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Y.C. is an Investigator of Howard Hughes Medical Institute. A.M. acknowledges support from the Pew Charitable Trusts, the Esther and A. & Joseph Klingenstein Fund and the Searle Scholars Program. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
Author information
Authors and Affiliations
Contributions
J.A.H. purified NK1R constructs, Gβ1γ2 and Nb35, established biochemical approaches to reconstitute a NK1R–miniG protein complex and generated all NK1R mutants. B.F. purified Gβ1γ2 and Nb35, prepared samples for cryo-EM, identified optimal freezing conditions for cryo-EM, screened samples by cryo-EM, collected cryo-EM data and determined high-resolution cryo-EM maps by extensive image processing under the guidance of A.M. and Y.C. J.A.H. built and refined models of NK1R–miniG protein complexes with input from B.F. and A.M. A.B.G. generated NK1R stable cell lines and performed cellular signaling experiments under the guidance of N.A.V. and D.M.T. M.A.D. and C.-M.S. performed and analyzed molecular dynamics simulations under the guidance of R.O.D. The paper was written by J.A.H., B.F., A.B.G., M.A.D., C.-M.S., R.O.D. and A.M., with edits from N.A.V., D.M.T. and Y.C. and with approval from all authors. The overall project was supervised by A.M.
Corresponding authors
Ethics declarations
Competing interests
Research in N.A.V.’s laboratory is funded, in part, by Takeda Pharmaceuticals and Endosome Therapeutics.
Additional information
Peer review information Nature Chemical Biology thanks Tao Che, Daniel Wacker and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
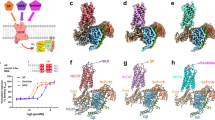
Extended Data Fig. 1 Biochemistry of active-state NK1R–miniG protein complexes.
(a) Size-exclusion chromatography of SP-bound NK1R, NK1R-miniGs/q70 and NK1R-miniGs399 shows an increase in the fraction of monomeric receptor species for NK1R-miniG fusion proteins. Size-exclusion chromatography traces and Coomassie stained SDS-PAGE gels of purified (b) SP-bound NK1R-miniGs/q70 complex, (c) SP-bound NK1R-miniGs399 complex, and (d) SP6-11-bound NK1R-miniGs/q70 complex. Uncropped versions of Coomassie stained SDS-PAGE gels are provided in Supplementary Fig. 1.
Extended Data Fig. 2 CryoEM data processing workflow for SP–NK1R-miniGs/q70 heterotrimeric complex.
(a) Representative micrograph of 3555 collected micrographs. Scale bar, 50 nm. (b) 2D-class averages for SP-NK1R-miniGs/q70 complex. (c) A flowchart representation of the processing pipeline used for structural determination of the SP-NK1R-miniGs/q70 complex. Contrast transfer function (CTF) estimation, 2D classification and all 3D classification jobs with alignment were performed with cryoSPARC. 3D classification without alignment was performed with RELION using a mask encompassing only the receptor transmembrane and final focused refinements were performed with cisTEM. Focused refinement masks are shown as red mesh. Gold-standard fourier shell correlation (GS-FSC) was calculated from a cryoSPARC Local Resolution job using the focused refinement mask encompassing the entire SP-NK1R-miniGs/q70 complex. A viewing distribution plot was generated using scripts from the pyEM software suite and visualized in ChimeraX. Directional FSC curves (dFSCs) are shown as purple lines and were determined as previously described in Dang, S. et al. Nature 552, 426-429 (2017).
Extended Data Fig. 3 Cryo-EM density map for NK1R-miniGs/q70 heterotrimeric complex.
(a) Unsharpened Cryo-EM density map for individual NK1R helices and Substance P density as determined by extending a 2.5 Å radius away from each modeled atom. Local resolution estimation of unsharpened Cryo-EM density maps for (b) SP-NK1R-miniGs/q70 and (c) SP6-11-NK1R-miniGs/q70 heterotrimeric complex from cryoSPARC. SP and SP6-11 density are highlighted in and shown at equivalent enclosed volume thresholds.
Extended Data Fig. 4 Structural hallmarks of NK1R activation.
(a) Alignment of the SP-NK1R-miniGs/q70 structure with an inactive-state NK1R structure (PDB: 6HLP24) reveals rearrangement of NK1R structural motifs indicative of class A GPCR activation, including: (b) displacement of the W6.48 ‘toggle-switch’ and (c) rearrangement of the ‘P5.50I3.40F6.44’ connector motif. (d) The non-canonical E782.50-N3017.49 interaction in NK1R is unchanged between inactive- and active-state structures. We compared the NK1R E782.50-N3017.49 interaction to the D2.50-N7.49 interaction in three class A neuropeptide-binding GPCRs, including: (e) the μ-opioid receptor (Active PDB: 5C1M17, Inactive PDB: 4DKL18), (f) the neurotensin 1 receptor (Active PDB: 6OS922, Inactive PDB: 4BUO23), and (g) the orexin 2 receptor (Active PDB: 7L1U, Inactive PDB: 5WQC). Alignment of the SP-NK1R-miniGs/q70 structure with (h) canonical (PDB: 6OS922) and (i) ‘non-canonical’ (PDB: 6OSA22) active-state NTS1R reveals that the miniGs/q70 protein adopts the canonical G protein coupling orientation.
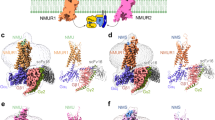
Extended Data Fig. 5 Comparison of SP-NK1R binding site to related Neuropeptide GPCRs and Inactive-State NK1R Structures.
Comparison of SP-bound NK1R-miniGs/q70 structure to neuropeptide GPCRs bound to peptidergic ligands, including: (a) the neurotensin 1 receptor bound to neurotensin 8-13 (PDB: 6OS922), (b) the μ-opioid receptor bound to the peptide mimetic agonist DAMGO (PDB: 6DDE26), and (c) the orexin 2 receptor bound to orexin B (PDB: 7L1U27). Alignment of SP-bound NK1R with inactive-state NK1R structures, including: (d) netupitant-bound (PDB: 6HLP24), (e) aprepitant-bound (PDB: 6HLO24), (f) L-760,735-bound (PDB: 6E5928), and (g) CP-99,994-bound NK1R (PDB: 6HLL24). (h) Antagonist chemical structures shown with regions that compete with SP binding site in red.
Extended Data Fig. 6 Signaling studies for NK1R mutations in the deep 7TM region.
Ca2+mobilization of wild-type and NK1R mutants after stimulation with (a) SP and (b) SP6-11. Signaling graphs represent the global fit of grouped data ± s.e.m. from at least three independent biological replicates. SP: N85D, n = 4; N85Q, n = 5; N89D, n = 5; H108A, n = 5; H108Q, n = 5; Y287F, n = 5; Y287H, n = 5. SP6-11: N85D, n = 3; N85Q, n = 4; N89D, n = 4; H108A, n = 4; H108Q, n = 4; Y287F, n = 4; Y287H, n = 3. Full quantitative parameters from this experiment are listed in Supplementary Table 1. (c) Cell-surface expression of deep 7TM NK1R mutants as determined by ELISA. Untransfected (UT) control shows low ELISA signal. Bar graphs represent mean ± s.e.m. from n = 4 independent biological replicates. Representative kinetic traces of (d) SP (e) NKA, and (f) SP6-11 elicited Ca2+ mobilization and cAMP accumulation from at least three independent biological replicates. Ca2+ signaling: SP, n = 10; NKA, n = 6; SP6-11, n = 4. IP1 accumulation: SP, n = 5; NKA, n = 3; SP6-11, n = 4. cAMP accumulation: SP, n = 12, NKA, n = 7, SP6-11, n = 4. Full quantitative parameters from this experiment are listed in Supplementary Table 2. (g) Ligand induced coupling of miniGs/q-Venus to hNK1R-RLuc as determined by BRET. Graphs represent the global fit of grouped data ± s.e.m. from n = 3 independent biological replicates. Full quantitative parameters from this experiment are listed in Supplementary Table 3. (h) Representative kinetic traces of SP, NKA, and SP6-11 induced recruitment of miniGs/q-Venus to hNK1R-RLuc as determined by BRET from n = 3 independent biological replicates. (i) Ligand induced coupling of miniGs-Venus to hNK1R-RLuc as determined by BRET. Graphs represent the global fit of grouped data ± s.e.m. from n = 3 independent biological replicates. Full quantitative parameters from this experiment are listed in Supplementary Table 3. (j) Representative kinetic traces of SP, NKA, and SP6-11 induced recruitment of miniGs-Venus to hNK1R-RLuc as determined by BRET from n = 3 independent biological replicates.
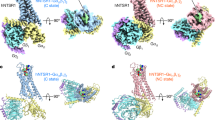
Extended Data Fig. 7 CryoEM data processing for SP-NK1R-miniGs399 complex.
(a) Representative micrograph of 3670 collected micrographs. Scale bar, 50 nm. (b) 2D-class averages for SP-NK1R-miniGs399 complex. (c) A flowchart representation of the processing pipeline used for structural determination of the SP-NK1R-miniGs399 complex. CTF Estimation, 2D classification and all 3D classification jobs with alignment were performed with cryoSPARC. 3D classification without alignment was performed with RELION using a mask encompassing only the receptor transmembrane and final focused refinements were performed with cisTEM. Focused refinement masks are shown as red mesh. Gold-standard fourier shell correlation (GS-FSC) was calculated from a cryoSPARC Local Resolution job using the focused refinement mask encompassing the entire SP-NK1R-miniGs399 complex. A viewing distribution plot was generated using scripts from the pyEM software suite and visualized in ChimeraX. Directional FSC (dFSC) are shown as purple lines and were determined as previously described in Dang, S. et al. Nature 552, 426-429 (2017).
Extended Data Fig. 8 CryoEM data processing for SP6-11-NK1R-miniGs/q70 complex.
(a) Representative micrograph of 3878 collected micrographs. Scale bar, 50 nm. (b) 2D-class averages for SP6-11-NK1R-miniGs/q70 complex. (c) A flowchart representation of the processing pipeline used for structural determination of the SP6-11-NK1R-miniGs/q70 complex. CTF Estimation, 2D classification and all 3D classification jobs with alignment were performed with cryoSPARC. 3D classification without alignment was performed with RELION using a mask encompassing only the receptor transmembrane and final focused refinements were performed with cisTEM. Focused refinement masks are shown as red mesh. GS-FSC was calculated from a cryoSPARC Local Resolution job using the focused refinement mask encompassing the entire SP6-11-NK1R-miniGs/q70 complex. A viewing distribution plot was generated using scripts from the pyEM software suite and visualized in ChimeraX. Directional FSC curves (dFSC) are shown in purple lines and were determined as previously described in Dang, S. et al. Nature 552, 426-429 (2017).
Extended Data Fig. 9 Comparison of NK1R G protein-complexes.
Alignment of SP-NK1R-miniGs/q70 and SP6-11-NK1R-miniGs/q70 through NK1R 7TM domain reveals minimal changes in (a) overall 7TM architecture, (b) overall peptide binding poses, and (c) insertion of miniG protein α5 helix in NK1R core. Alignment of SP-NK1R-miniGs/q70 and SP-NK1R-miniGs399 through NK1R 7TM domain reveals minimal changes in (d) overall 7TM architecture, (e) overall SP binding pose, and (f) insertion of miniG protein α5 helix in NK1R core.
Extended Data Fig. 10 Signaling studies for NK1R ECL2 mutations.
(a) Fraction of time R177 is in contact with SP vs. SP6-11 in molecular dynamics simulations. Bar graphs show mean ± s.e.m. from twelve independent molecular dynamics simulations under each condition. SP spent more time in contact with R177 than SP6-11 (p < 0.05, two-sided Welch’s t-test; see Methods). Our simulations are not sufficiently long to guarantee convergence of this quantity. (b) Ca2+ mobilization and (c) cAMP accumulation of wild-type and ECL2 NK1R mutants after stimulation with SP. Signaling graphs represent the global fit of grouped data ± s.e.m. from at least three independent biological replicates. Ca2+ signaling: WT, n = 9; M174V, n = 5; R177K, n = 6; M181F, n = 5; M181V, n = 5. cAMP accumulation: WT, n = 12; M174V, n = 3; R177K, n = 4; M181F, n = 3; M181V, n = 4. Full quantitative parameters from this experiment are listed in Supplementary Table 1. (d) Cell-surface expression of ECL2 NK1R mutants as determined by ELISA. Untransfected (UT) control shows low ELISA signal. Bar graphs represent mean ± s.e.m. from n = 4 independent biological replicates. (e) Ca2+ mobilization and cAMP accumulation of wild-type, M174I, and R177M NK1R mutants after stimulation with SP6-11. Signaling graphs represent the global fit of grouped data ± s.e.m. from at lease three independent biological replicates. Ca2+ signaling: WT, n = 8; M174I, n = 4; R177M, n = 4. cAMP accumulation: WT, n = 11; M174I, n = 3; R177M, n = 4. Full quantitative parameters from this experiment are listed in Supplementary Table 1. Representative kinetic traces of SP-induced Ca2+ mobilization and cAMP accumulation for (f) NK1R M174I and (g) NK1R R177M from at least three independent biological replicates. Ca2+ signaling: WT, n = 9; M174I, n = 5; R177M, n = 9. cAMP accumulation: WT, n = 12, M174I, n = 3, R177M, n = 8. Full quantitative parameters from this experiment are listed in Supplementary Table 1.
Supplementary information
Supplementary Information
Supplementary Tables 1–4 and Fig. 1.
Source data
Source Data Fig. 1
Uncropped Coomassie-stained SDS–PAGE gels from Extended Data Fig. 1.
Rights and permissions
About this article
Cite this article
Harris, J.A., Faust, B., Gondin, A.B. et al. Selective G protein signaling driven by substance P–neurokinin receptor dynamics. Nat Chem Biol 18, 109–115 (2022). https://doi.org/10.1038/s41589-021-00890-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00890-8
This article is cited by
-
Improving resolution and resolvability of single-particle cryoEM structures using Gaussian mixture models
Nature Methods (2024)
-
Fluorine-19 labeling of the tryptophan residues in the G protein-coupled receptor NK1R using the 5-fluoroindole precursor in Pichia pastoris expression
Journal of Biomolecular NMR (2024)
-
Molecular basis for the selective G protein signaling of somatostatin receptors
Nature Chemical Biology (2023)
-
The role of G protein conformation in receptor–G protein selectivity
Nature Chemical Biology (2023)
-
Specific binding of GPR174 by endogenous lysophosphatidylserine leads to high constitutive Gs signaling
Nature Communications (2023)