Abstract
Multiple Ras proteins, including N-Ras, depend on a palmitoylation/depalmitoylation cycle to regulate their subcellular trafficking and oncogenicity. General lipase inhibitors such as Palmostatin M (Palm M) block N-Ras depalmitoylation, but lack specificity and target several enzymes displaying depalmitoylase activity. Here, we describe ABD957, a potent and selective covalent inhibitor of the ABHD17 family of depalmitoylases, and show that this compound impairs N-Ras depalmitoylation in human acute myeloid leukemia (AML) cells. ABD957 produced partial effects on N-Ras palmitoylation compared with Palm M, but was much more selective across the proteome, reflecting a plasma membrane-delineated action on dynamically palmitoylated proteins. Finally, ABD957 impaired N-Ras signaling and the growth of NRAS-mutant AML cells in a manner that synergizes with MAP kinase kinase (MEK) inhibition. Our findings uncover a surprisingly restricted role for ABHD17 enzymes as regulators of the N-Ras palmitoylation cycle and suggest that ABHD17 inhibitors may have value as targeted therapies for NRAS-mutant cancers.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All mass spectrometry data are available via ProteomeXchange with identifier PXD023932. Source data are provided with this paper. All other data that support the findings of this study are available from the corresponding authors upon reasonable request.
Code availability
Code used to process mass spectrometric and imaging data are available on GitHub: https://github.com/cravattlab/remsberg_ncb_2021.
References
Schubbert, S., Shannon, K. & Bollag, G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer 7, 295–308 (2007).
Ostrem, J. M., Peters, U., Sos, M. L., Wells, J. A. & Shokat, K. M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013).
Canon, J. et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019).
Omerovic, J., Laude, A. J. & Prior, I. A. Ras proteins: paradigms for compartmentalised and isoform-specific signalling. Cell. Mol. Life Sci. 64, 2575–2589 (2007).
Hancock, J. F., Paterson, H. & Marshall, C. J. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63, 133–139 (1990).
Rocks, O. et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307, 1746–1752 (2005).
Dekker, F. J. et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat. Chem. Biol. 6, 449–456 (2010).
Hedberg, C. et al. Development of highly potent inhibitors of the Ras-targeting human acyl protein thioesterases based on substrate similarity design. Angew. Chem. Int. Ed. Engl. 50, 9832–9837 (2011).
Martin, B. R., Wang, C., Adibekian, A., Tully, S. E. & Cravatt, B. F. Global profiling of dynamic protein palmitoylation. Nat. Methods 9, 84–89 (2011).
Duncan, J. A. & Gilman, A. G. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J. Biol. Chem. 273, 15830–15837 (1998).
Rusch, M. et al. Identification of acyl protein thioesterases 1 and 2 as the cellular targets of the Ras-signaling modulators palmostatin B and M. Angew. Chem. Int. Ed. Engl. 50, 9838–9842 (2011).
Lin, D. T. & Conibear, E. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. eLife 4, e11306 (2015).
Martin, B. R. & Cravatt, B. F. Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods 6, 135–138 (2009).
Yokoi, N. et al. Identification of PSD-95 depalmitoylating enzymes. J. Neurosci. 36, 6431–6444 (2016).
Jia, L. et al. A mechanism regulating G protein-coupled receptor signaling that requires cycles of protein palmitoylation and depalmitoylation. J. Biol. Chem. 289, 6249–6257 (2014).
Won, S. J. & Martin, B. R. Temporal profiling establishes a dynamic S-palmitoylation cycle. ACS Chem. Biol. 13, 1560–1568 (2018).
Zhang, M. M., Tsou, L. K., Charron, G., Raghavan, A. S. & Hang, H. C. Tandem fluorescence imaging of dynamic S-acylation and protein turnover. Proc. Natl Acad. Sci. USA 107, 8627–8632 (2010).
Cao, Y. et al. ABHD10 is an S-depalmitoylase affecting redox homeostasis through peroxiredoxin-5. Nat. Chem. Biol. 15, 1232–1240 (2019).
Adibekian, A. et al. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat. Chem. Biol. 7, 469–478 (2011).
Chang, J. W., Nomura, D. K. & Cravatt, B. F. A potent and selective inhibitor of KIAA1363/AADACL1 that impairs prostate cancer pathogenesis. Chem. Biol. 18, 476–484 (2011).
Hsu, K. L. et al. DAGLbeta inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nat. Chem. Biol. 8, 999–1007 (2012).
Niphakis, M. J. & Cravatt, B. F. Enzyme inhibitor discovery by activity-based protein profiling. Annu. Rev. Biochem. 83, 341–377 (2014).
Otrubova, K., Chatterjee, S., Ghimire, S., Cravatt, B. F. & Boger, D. L. N-Acyl pyrazoles: effective and tunable inhibitors of serine hydrolases. Bioorg. Med. Chem. 27, 1693–1703 (2019).
Liu, Y., Patricelli, M. P. & Cravatt, B. F. Activity-based protein profiling: the serine hydrolases. Proc. Natl Acad. Sci. USA 96, 14694–14699 (1999).
Cognetta, A. B. 3rd et al. Selective N-hydroxyhydantoin carbamate inhibitors of mammalian serine hydrolases. Chem. Biol. 22, 928–937 (2015).
Zambetti, N. A. et al. Genetic disruption of N-RasG12D palmitoylation perturbs hematopoiesis and prevents myeloid transformation in mice. Blood 135, 1772–1782 (2020).
Charron, G. et al. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J. Am. Chem. Soc. 131, 4967–4975 (2009).
Rostovtsev, V. V., Green, L. G., Fokin, V. V. & Sharpless, K. B. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective ‘ligation’ of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 41, 2596–2599 (2002).
Drisdel, R. C. & Green, W. N. Labeling and quantifying sites of protein palmitoylation. Biotechniques 36, 276–285 (2004).
Zecha, J. et al. Peptide level turnover measurements enable the study of proteoform dynamics. Mol. Cell. Proteomics 17, 974–992 (2018).
Chen, B. et al. ZDHHC7-mediated S-palmitoylation of scribble regulates cell polarity. Nat. Chem. Biol. 12, 686–693 (2016).
Kamijo, A., Saitoh, Y., Ohno, N., Ohno, S. & Terada, N. Immunohistochemical study of the membrane skeletal protein, membrane protein palmitoylated 6 (MPP6), in the mouse small intestine. Histochem. Cell Biol. 145, 81–92 (2016).
Jones, T. L. & Gutkind, J. S. Galpha12 requires acylation for its transforming activity. Biochemistry 37, 3196–3202 (1998).
Saraceno, C. et al. SAP97-mediated ADAM10 trafficking from Golgi outposts depends on PKC phosphorylation. Cell Death Dis. 5, e1547 (2014).
Choi, S. I., Vidal, R., Frangione, B. & Levy, E. Axonal transport of British and Danish amyloid peptides via secretory vesicles. FASEB J. 18, 373–375 (2004).
Xu, J. et al. Inhibiting the palmitoylation/depalmitoylation cycle selectively reduces the growth of hematopoietic cells expressing oncogenic Nras. Blood 119, 1032–1035 (2012).
Zhao, W. et al. A new bliss independence model to analyze drug combination data. J. Biomol. Screen. 19, 817–821 (2014).
Ahearn, I. M., Haigis, K., Bar-Sagi, D. & Philips, M. R. Regulating the regulator: post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 13, 39–51 (2011).
Ryan, M. B. & Corcoran, R. B. Therapeutic strategies to target RAS-mutant cancers. Nat. Rev. Clin. Oncol. 15, 709–720 (2018).
Cox, A. D., Der, C. J. & Philips, M. R. Targeting RAS membrane association: back to the future for anti-RAS drug discovery? Clin. Cancer Res. 21, 1819–1827 (2015).
Cox, A. D., Fesik, S. W., Kimmelman, A. C., Luo, J. & Der, C. J. Drugging the undruggable RAS: mission possible? Nat. Rev. Drug Discov. 13, 828–851 (2014).
Hernandez, J. L. et al. APT2 inhibition restores scribble localization and S-palmitoylation in Snail-transformed cells. Cell Chem. Biol. 24, 87–97 (2017).
Vartak, N. et al. The autodepalmitoylating activity of APT maintains the spatial organization of palmitoylated membrane proteins. Biophys. J. 106, 93–105 (2014).
Kathayat, R. S. et al. Active and dynamic mitochondrial S-depalmitoylation revealed by targeted fluorescent probes. Nat. Commun. 9, 334 (2018).
Levental, I., Lingwood, D., Grzybek, M., Coskun, U. & Simons, K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc. Natl Acad. Sci. USA 107, 22050–22054 (2010).
Chandra, A. et al. The GDI-like solubilizing factor PDEdelta sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 14, 148–158 (2011).
Zhou, M. et al. VPS35 binds farnesylated N-Ras in the cytosol to regulate N-Ras trafficking. J. Cell Biol. 214, 445–458 (2016).
Nakai, K. & Horton, P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–36 (1999).
Patricelli, M. P., Giang, D. K., Stamp, L. M. & Burbaum, J. J. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics 1, 1067–1071 (2001).
Chang, J. W., Cognetta, A. B. 3rd, Niphakis, M. J. & Cravatt, B. F. Proteome-wide reactivity profiling identifies diverse carbamate chemotypes tuned for serine hydrolase inhibition. ACS Chem. Biol. 8, 1590–1599 (2013).
Hatfield, M. J. et al. Biochemical and molecular analysis of carboxylesterase-mediated hydrolysis of cocaine and heroin. Br. J. Pharmacol. 160, 1916–1928 (2010).
Inloes, J. M. et al. The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc. Natl Acad. Sci. USA 111, 14924–14929 (2014).
Xu, T. et al. ProLuCID: an improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteomics 129, 16–24 (2015).
Cociorva, D., Tabb, D. L. & Yates, J. R. Validation of tandem mass spectrometry database search results using DTASelect.Curr. Protoc. Bioinformatics Chapter 13, Unit 13.4 (2007).
Zuber, J. et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 25, 1628–1640 (2011).
Burgess, M. R. et al. Preclinical efficacy of MEK inhibition in Nras-mutant AML. Blood 124, 3947–3955 (2014).
Wang, Y. et al. Expedited mapping of the ligandable proteome using fully functionalized enantiomeric probe pairs. Nat. Chem. 11, 1113–1123 (2019).
Vinogradova, E. V. et al. An activity-guided map of electrophile–cysteine interactions in primary human T cells. Cell 182, 1009–1026 e29 (2020).
Adler, J. & Parmryd, I. Quantifying colocalization by correlation: the pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytometry A 77, 733–742 (2010).
Acknowledgements
This work was supported by grants from the NIH (CA193994, CA231991 and CA72614), an American Cancer Society postdoctoral fellowship PF-18-217-01-CDD (to J.R.R.), the Leukemia and Lymphoma Society (LLS Fellowship 5465-18 to N.A.Z.), and the Damon Runyon Cancer Research Foundation (Fellowship DRG-2149-13 to A.J.F). We thank S. Henderson for assistance with the microscopy studies, and T. Jung and R. Park for assistance with processing the SILAC-TMT proteomic data.
Author information
Authors and Affiliations
Contributions
J.R.R., R.M.S., N.A.Z., M.J.N., K.S. and B.F.C. conceived the project and wrote the paper. J.R.R. and M.M.D. performed palmitoyl-proteomics experiments. M.J.N. led efforts to identify ABD957. R.M.S. performed N-Ras palmitoylation assays. N.A.Z., J.R.R., A.J.F., A.I., A.L., M.P. and B.H. performed proliferation and N-Ras signaling assays. T.W.H. performed imaging experiments. A.J.F. developed syngeneic AML cell lines. N.N., C.L.H. and R.M.S. performed gel- and MS-ABPP experiments. N.N. and C.L.H. performed compound characterization and biochemical assays for inhibitor characterization. K.M.L. and R.M.S. analyzed MS-ABPP experiments and analyzed spectra. S.K.R. synthesized Palmostatin M. A.R.H. provided synthetic expertise and contributed to related studies not detailed in this paper. J.R.R., R.M.S., N.A.Z., T.W.H., K.M.L., M.M.D. and M.J.N. performed data analysis and visualization.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemical Biology thanks Matthew Bogyo, Mark Philips and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of ABHD17 inhibitors.
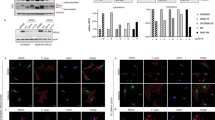
a, Representative gel-ABPP image showing FP-rhodamine labeling of recombinant human ABHD17B (hABHD17B) in proteomic lysates of stably transfected HEK293T cells. b, IC50 curves and representative gel-ABPP images of human ABHD17B and LYPLA1 activity in HEK293T cell lysate treated with compounds 5–8 and ABD957 for 30 min. Data represent average values ± s.d. (n = 3 independent experiments). c, MS-ABPP data of serine hydrolase activities in the particulate fraction of OCI-AML3 proteomes treated with compounds 5-8 and ABD957 for 30 min (10 µM). Data are from single experiments performed at the indicated concentrations for each compound. d, MS-ABPP data of serine hydrolase activities in OCI-AML3 cells treated in situ with Palm M (10 µM) or HDFP (20 µM) for 2 h. Data represent the median from three experiments corresponding to independent treatments of cells with compound (Palm M or HDFP) and error bars represent s.d.
Extended Data Fig. 2 Analysis of off-targets of ABD957 and control probes accounting for these serine hydrolases.
a, Structures of ABD298, an ABHD13 inhibitor, and JJH254, a previously reported LYPLA1/2 inhibitor25. b, MS-ABPP data of serine hydrolase activities in OCI-AML3 cells treated in situ with JJH254 (1 µM) or ABD298 (500 nM) for 2 h. Data represent the median from three experiments corresponding to independent treatments of cells with each compound (JJH254 or ABD298) and error bars represent standard deviation. c, IC50 curves and representative gel-ABPP images for ABD957 and ABD298 determined in proteomes of HEK293T cells expressing recombinant mouse ABHD13. Lysates were treated in vitro with inhibitor for 30 min at 37 oC, followed by incubation with FP-Rh for 30 min, RT. Experiments were performed in triplicate. Error bars represent s.d. and center around the mean. d, IC50 curve and representative gel-ABPP image for ABD957 determined for endogenous human ABHD6 in PC3 cell proteomes following a 30 min inhibitor treatment. Experiments were performed in triplicate. Error bars represent s.d. and center around the mean. e, IC50 curve for ABD957 derived by measuring the CES2-dependent rate of hydrolysis (background substracted) of the substrate 4-nitrophenyl acetate (pNPA) in proteomes of HEK293T cells expressing recombinant human CES2 following a 30 min inhibitor treatment. The mean reaction velocity (Vmean, ΔAbs / min·µg protein) in mock- versus CES2-transfected proteomes was 8.7 ± 1.9 min−1·µg−1 and 31 ± 1.2 min−1·µg−1, respectively. Data presented for all IC50 determinations represent average values ± s.d. (n = 3 independent experiments).
Extended Data Fig. 3 Schematic of a dynamic palmitoylation assay using metabolic labeling with 17-ODYA.
Black arrowheads next to mock gel mark proteins that show dynamic palmitoylation fully (one arrowhead), partially (two arrowheads), or not (three arrowheads) preserved by serine hydrolase inhibition.
Extended Data Fig. 4 Global palmitoylation effects of Palm M and ABD957 in leukemia cells.
a, MS-based proteomics in OCI-AML3 cells showing the 17- ODYA labeling at t0. The results indicate that Palm M increases the apparent palmitoylation state of several proteins prior to the chase period and, as shown in Fig. 3a, some of these proteins are not dynamically palmitoylated (blue proteins in Fig. 3a). b, Bar graph quantifying different categories of N-Ras peptides from MS-based proteomics experiments in ON cells. Data represent average values relative to DMSO t0 ± s.d. (n = 4 from two biological replicates). Statistical significance was calculated with unpaired two-tailed Student’s t-test with equal variance, ***P < 0.001, ****P < 0.0001 represent significant increase compared to DMSO t1 (also see Supplementary Dataset 3). P values were 5.5×10-5 (Palm M GFP specific), 1.8×10-4 (ABD957 GFP specific), 2.1×10-5 (Palm M N-Ras specific), 2.7×10-4 (ABD957 N-Ras specific). c, d, Scatter plots as described in Fig. 3a in ON cells.
Extended Data Fig. 5 Effects of ABD957 treatment on N-Ras turnover in OCI-AML3 cells.
a, Degradation and synthesis curves for N-Ras in OCI-AML3 cells treated with DMSO or ABD957 (500 nM). Each dot represents a peptide spectrum match (PSM) from one of three biological replicates. OCI-AML3 cells grown in light SILAC media were pelleted and resuspended in heavy SILAC media containing DMSO or ABD957 and harvested at the indicated times. Also see Supplementary Dataset 4.
Extended Data Fig. 6 Global palmitoylation effects of Palm M and ABD957 in NB-4 cells.
a, b, Bar graphs quantifying SCRIB and N-Ras palmitoylation from MS-based proteomic experiments in NB-4 cells. Data represent average values relative to DMSO t0 ± s.d. (biological replicates; n=2 for Palm M t1, n=8 for all other conditions). Statistical significance was calculated with unpaired two-tailed Student’s t-test with equal variance, *P < 0.05, ****P < 0.0001 represent significant increase compared to DMSO t1 (also see Supplementary Dataset 2 and 3). P values were 6.5×10-7 (SCRIB), 0.013 (N-Ras).
Extended Data Fig. 7 Quantification of data shown in Fig. 5f; average values ± s.d. (n = 4 independent experiments).
Statistical significance was calculated with unpaired two-tailed Student’s t-test with unequal variance compared to DMSO control, *P < 0.05, **P < 0.01, ****P < 0.0001. P values were 4.8×10-6 (Palm M OCI-AML3), 0.012 (ABD957 OCI-AML3), 0.0016 (Palm M ABHD17-DKO 1), 0.0014 (Palm M ABHD17-DKO 2), 2.2×10-5 (Palm M LYPLA-DKO 1), 0.0010 (ABD957 LYPLA-DKO 1), 7.4×10-5 (Palm M LYPLA-DKO 2), 2.1×10-5 (ABD957 LYPLA-DKO 2).
Extended Data Fig. 8 MS-based proteomic analysis of dynamic palmitoylation in parental and ABHD17- DKO cells.
a, Profiles for ABD957-regulated palmitoylated proteins from mass spectrometry (MS)-based experiments of OCI-AML3 and ABHD17-DKO 1 cells performed as described in Extended Data Fig. 3. Cells were preincubated with ABD957 (500 nM) or DMSO for 1h, metabolically labeled with 20 µM 17-ODYA for 1h (t0) and chased with media lacking 17-ODYA for 1 h (t1) containing ABD957 (500 nM) or DMSO control. Data represent average values relative to each cell lines corresponding DMSO t0 control ± s.d. (biological replicates, n = 4 for DMSO t0 values, n=5 for all others). Statistical significance was calculated with unpaired two-tailed Student’s t-test with equal variance, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, compared to DMSO t1 for left panels or compared to DMSO t1/t0 ratio between OCI-AML3 and ABHD17-DKO 1 cells for right panels. P values were 0.0096 (OCI-AML3 ABD957 N-Ras), 7.8×10−4 (OCI- AML3 DMSO t1/t0 N-Ras), 2.4×10−5 (OCI-AML3 ABD957 SCRIB), 4.4×10−4 (OCI-AML3 DMSO t1/t0 SCRIB), 1.9×10−6 (OCI-AML3 ABD957 MPP6), 1.2×10−8 (OCI-AML3 DMSO t1/t0 MPP6), 3.9×10−4 (OCI-AML3 ABD957 GNA12), 0.0044 (OCI-AML3 DMSO t1/t0 GNA12). b, Effect of ABD957 on dynamically palmitoylated proteins in OCI-AML3 and ABHD17-DKO 1 cells. Data represent average values ± s.d. (n = 5 biological replicates). Statistical significance was calculated for proteins with >2-fold increases in ABD957-treated OCI-AML3 cells with unpaired two-tailed Student’s t-test with equal variance, **P < 0.01, ****P < 0.0001 compared to ABHD17-DKO 1 cells. P values were 0.0049 (N-Ras), 2.3×10-7 (SCRIB), 7.6×10-8 (MPP6), 8.1×10-5 (GNA12).
Supplementary information
Supplementary Information
Supplementary Tables 1–3, Figs. 1–5, Video 1 caption and Note - Chemistry Methods, NMR spectra.
Supplementary Dataset 1
MS-ABPP studies for target profiling.
Supplementary Dataset 2
Data from mass spectrometry-based studies of hydroxylamine-sensitive 17-ODYA-labeled proteins.
Supplementary Dataset 3
Data from mass spectrometry-based studies of dynamic palmitoylation.
Supplementary Dataset 4
Data from SILAC-TMT mass spectrometry studies to assess protein turnover.
Supplementary Video 1
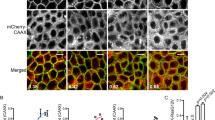
Effect of inhibitor treatment on GFP–N-Ras localization in ON cells. Representative time-lapse videos from ON cells before and after treatment with Palm M (10 µM) (a), ABD957 (500 nM) (b), or DMSO control (c). Green channel shows GFP–N-Ras signal. Frames were acquired every 90 s over 680 x 680 pixels for 10 frames prior to compound infusion and a total of 35 frames is shown, with each frame requiring 13.37 s to scan. Relative acquisition time is shown in red text in the upper left. Frames where compound infusion begins are captioned in the upper left in red text with the name of compound used for the experiment. Data shown are representative of three biological replicates. One medial z-plane over a 400 x 400-pixel area is shown for clarity. Scale bar, 10 μm.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots and/or gels.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
About this article
Cite this article
Remsberg, J.R., Suciu, R.M., Zambetti, N.A. et al. ABHD17 regulation of plasma membrane palmitoylation and N-Ras-dependent cancer growth. Nat Chem Biol 17, 856–864 (2021). https://doi.org/10.1038/s41589-021-00785-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00785-8
This article is cited by
-
Protein lipidation in health and disease: molecular basis, physiological function and pathological implication
Signal Transduction and Targeted Therapy (2024)
-
Mechanisms and functions of protein S-acylation
Nature Reviews Molecular Cell Biology (2024)
-
Protein lipidation in cancer: mechanisms, dysregulation and emerging drug targets
Nature Reviews Cancer (2024)
-
Regulation of RAS palmitoyltransferases by accessory proteins and palmitoylation
Nature Structural & Molecular Biology (2024)
-
Palmitoylation landscapes across human cancers reveal a role of palmitoylation in tumorigenesis
Journal of Translational Medicine (2023)