Abstract
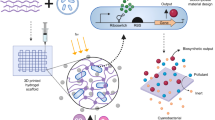
Genetically modified microorganisms (GMMs) can enable a wide range of important applications including environmental sensing and responsive engineered living materials. However, containment of GMMs to prevent environmental escape and satisfy regulatory requirements is a bottleneck for real-world use. While current biochemical strategies restrict unwanted growth of GMMs in the environment, there is a need for deployable physical containment technologies to achieve redundant, multi-layered and robust containment. We developed a hydrogel-based encapsulation system that incorporates a biocompatible multilayer tough shell and an alginate-based core. This deployable physical containment strategy (DEPCOS) allows no detectable GMM escape, bacteria to be protected against environmental insults including antibiotics and low pH, controllable lifespan and easy retrieval of genomically recoded bacteria. To highlight the versatility of DEPCOS, we demonstrated that robustly encapsulated cells can execute useful functions, including performing cell–cell communication with other encapsulated bacteria and sensing heavy metals in water samples from the Charles River.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data supporting this study are presented in the main text and Supplementary Information, and are available from the corresponding authors upon request. Source data are provided with this paper.
References
Singh, J. S., Abhilash, P. C., Singh, H. B., Singh, R. P. & Singh, D. P. Genetically engineered bacteria: an emerging tool for environmental remediation and future research perspectives. Gene 480, 1–9 (2011).
Farrar, K., Bryant, D. & Cope-Selby, N. Understanding and engineering beneficial plant-microbe interactions: plant growth promotion in energy crops. Plant Biotechnol. J. 12, 1193–1206 (2014).
Tang, T.-C. et al. Materials design by synthetic biology. Nat. Rev. Mater. https://doi.org/10.1038/s41578-020-00265-w (2020).
Dana, G. V., Kuiken, T., Rejeski, D. & Snow, A. A. Synthetic biology: four steps to avoid a synthetic-biology disaster. Nature 483, 29 (2012).
Torres, L., Krüger, A., Csibra, E., Gianni, E. & Pinheiro, V. B. Synthetic biology approaches to biological containment: pre-emptively tackling potential risks. Essays Biochem. 60, 393–410 (2016).
Epstein, M. M. & Vermeire, T. Scientific opinion on risk assessment of synthetic biology. Trends Biotechnol. 34, 601–603 (2016).
Wright, O., Stan, G. B. & Ellis, T. Building-in biosafety for synthetic biology. Microbiol. 159, 1221–1235 (2013).
Lee, J. W., Chan, C. T. Y., Slomovic, S. & Collins, J. J. Next-generation biocontainment systems for engineered organisms. Nat. Chem. Biol. 14, 530–537 (2018).
Chan, C. T. Y., Lee, J. W., Cameron, D. E., Bashor, C. J. & Collins, J. J. ‘Deadman’ and ‘Passcode’ microbial kill switches for bacterial containment. Nat. Chem. Biol. 12, 82–86 (2016).
Wright, O., Delmans, M., Stan, G. B. & Ellis, T. GeneGuard: a modular plasmid system designed for biosafety. ACS Synth. Biol. 4, 307–316 (2015).
Gallagher, R. R., Patel, J. R., Interiano, A. L., Rovner, A. J. & Isaacs, F. J. Multilayered genetic safeguards limit growth of microorganisms to defined environments. Nucleic Acids Res. 43, 1945–1954 (2015).
Rovner, A. J. et al. Recoded organisms engineered to depend on synthetic amino acids. Nature 518, 89–93 (2015).
Hoffman, A. S. Hydrogels for biomedical applications. Adv. Drug Deliv. Rev. 64, 18–23 (2012).
Choi, M. et al. Light-guiding hydrogels for cell-based sensing and optogenetic synthesis in vivo. Nat. Photonics 7, 987–994 (2013).
Anselmo, A. C., McHugh, K. J., Webster, J., Langer, R. & Jaklenec, A. Layer-by-layer encapsulation of probiotics for delivery to the microbiome. Adv. Mater. 28, 9486–9490 (2016).
Lee, K. Y. & Mooney, D. J. Alginate: properties and biomedical applications. Prog. Polym. Sci. 37, 106–126 (2012).
Kearney, C. J. & Mooney, D. J. Macroscale delivery systems for molecular and cellular payloads. Nat. Mater. 12, 1004–1017 (2013).
Billiet, T., Vandenhaute, M., Schelfhout, J., Van Vlierberghe, S. & Dubruel, P. A review of trends and limitations in hydrogel-rapid prototyping for tissue engineering. Biomaterials 33, 6020–6041 (2012).
Kim, B. J. et al. Cytoprotective alginate/polydopamine core/shell microcapsules in microbial encapsulation. Angew. Chem. Int. Ed. 53, 14443–14446 (2014).
Li, P., Müller, M., Chang, M. W., Frettlöh, M. & Schönherr, H. Encapsulation of autoinducer sensing reporter bacteria in reinforced alginate-based microbeads. ACS Appl. Mater. Interfaces 9, 22321–22331 (2017).
Zhang, B. B., Wang, L., Charles, V., Rooke, J. C. & Su, B. L. Robust and biocompatible hybrid matrix with controllable permeability for microalgae encapsulation. ACS Appl. Mater. Interfaces 8, 8939–8946 (2016).
Sun, J. Y. et al. Highly stretchable and tough hydrogels. Nature 489, 133–136 (2012).
Liu, X. et al. Stretchable living materials and devices with hydrogel-elastomer hybrids hosting programmed cells. Proc. Natl Acad. Sci. USA 114, 2200–2205 (2017).
Valade, D., Wong, L. K., Jeon, Y., Jia, Z. & Monteiro, M. J. Polyacrylamide hydrogel membranes with controlled pore sizes. J. Polym. Sci., Part A: Polym. Chem. 51, 129–138 (2013).
Atkinson, J. The Mechanics of Soils and Foundations 2nd edn (CRC Press, 2007); https://doi.org/10.1201/9781315273549
Liu, X. et al. Ingestible hydrogel device. Nat. Commun. 10, 493 (2019).
Houghton, L. A. et al. Motor activity of the gastric antrum, pylorus, and duodenum under fasted conditions and after a liquid meal. Gastroenterology 94, 1276–1284 (1988).
Zarket, B. C. & Raghavan, S. R. Onion-like multilayered polymer capsules synthesized by a bioinspired inside-out technique. Nat. Commun. 8, 193 (2017).
Eun, Y. J., Utada, A. S., Copeland, M. F., Takeuchi, S. & Weibel, D. B. Encapsulating bacteria in agarose microparticles using microfluidics for high-throughput cell analysis and isolation. ACS Chem. Biol. 6, 260–266 (2011).
Kong, H. J., Kim, E. S., Huang, Y. C. & Mooney, D. J. Design of biodegradable hydrogel for the local and sustained delivery of angiogenic plasmid DNA. Pharm. Res. 25, 1230–1238 (2008).
Farzadfard, F. & Lu, T. K. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346, 1256272–1256272 (2014).
Golmohamadi, M. & Wilkinson, K. J. Diffusion of ions in a calcium alginate hydrogel-structure is the primary factor controlling diffusion. Carbohydr. Polym. 94, 82–87 (2013).
Li, Z. et al. Biofilm-inspired encapsulation of probiotics for the treatment of complex infections. Adv. Mater. 30, e1803925 (2018).
Bjerketorp, J., Håkansson, S., Belkin, S. & Jansson, J. K. Advances in preservation methods: keeping biosensor microorganisms alive and active. Curr. Opin. Biotechnol. 17, 43–49 (2006).
Roggo, C. & van der Meer, J. R. Miniaturized and integrated whole cell living bacterial sensors in field applicable autonomous devices. Curr. Opin. Biotechnol. 45, 24–33 (2017).
Kim, B. C. & Gu, M. B. A bioluminescent sensor for high throughput toxicity classification. Biosens. Bioelectron. 18, 1015–1021 (2003).
Cevenini, L., Calabretta, M. M., Tarantino, G., Michelini, E. & Roda, A. Smartphone-interfaced 3D printed toxicity biosensor integrating bioluminescent ‘sentinel cells’. Sens. Actuators, B Chem. 225, 249–257 (2016).
Mimee, M. et al. An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 360, 915–918 (2018).
Pedersen, M. W. et al. Ancient and modern environmental DNA. Philos. Trans. R. Soc. B. Biol. Sci. 370, 20130383 (2015).
Sheth, R. U. & Wang, H. H. DNA-based memory devices for recording cellular events. Nat. Rev. Genet. 19, 718–732 (2018).
Farzadfard, F., Gharaei, N., Citorik, R. J. & Lu, T. K. Efficient retroelement-mediated DNA writing in bacteria. Preprint at bioRxiv https://doi.org/10.1101/2020.02.21.958983 (2020).
Weiss, R. & Knight, T. F. Jr. in Cellular Computing (ed. Amos, M.) 120–121 (Oxford Univ. Press, 2004); https://doi.org/10.1093/oso/9780195155396.003.0012
Chen, M. T. & Weiss, R. Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat. Biotechnol. 23, 1551–1555 (2005).
Järup, L. & Åkesson, A. Current status of cadmium as an environmental health problem. Toxicol. Appl. Pharmacol. 238, 201–208 (2009).
Brocklehurst, K. R. et al. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol. 31, 893–902 (1999).
Knierim, C., Greenblatt, C. L., Agarwal, S. & Greiner, A. Blocked bacteria escape by ATRP grafting of a PMMA shell on PVA microparticles. Macromol. Biosci. 14, 537–545 (2014).
Belkin, S. et al. Remote detection of buried landmines using a bacterial sensor. Nat. Biotechnol. 35, 308–310 (2017).
De Las Heras, A., Carreño, C. A. & De Lorenzo, V. Stable implantation of orthogonal sensor circuits in Gram-negative bacteria for environmental release. Environ. Microbiol. 10, 3305–3316 (2008).
Tomović, N. S., Trifković, K. T., Rakin, M. P., Rakin, M. B. & Bugarski, B. M. Influence of compression speed and deformation percentage on mechanical properties of calcium alginate particles. Chem. Ind. Chem. Eng. Q. 21, 411–417 (2015).
Acknowledgements
We thank F. Farzadfard for proving the SCRIBE strains and M. Mimee for providing the heme sensing strain. We thank S. Lin, N. Roquet, R. Citorik and S. Lemire for useful discussions. T.K.L. is grateful for funding received from the National Institutes of Health (NIH) New Innovator Award (no. 1DP2OD008435), NIH National Centers for Systems Biology (no. 1P50GM098792), the US Office of Naval Research (no. N00014-13-1-0424) and the Defense Advanced Research Projects Agency (no. HR0011-15-C-0091). X.Z. is grateful for funding received from the NIH (no. 1R01HL153857-01), the National Science Foundation (no. EFMA-1935291) and the US Army Research Office through the Institute for Soldier Nanotechnologies at MIT (no. W911NF-13-D-0001). C.F.-N. holds a Presidential Professorship at the University of Pennsylvania, is a recipient of the Langer Prize by the AIChE Foundation and acknowledges funding from the Institute for Diabetes, Obesity, and Metabolism, the Penn Mental Health AIDS Research Center of the University of Pennsylvania, the National Institute of General Medical Sciences of the NIH (no. R35GM138201), and the Defense Threat Reduction Agency (no. HDTRA11810041 and HDTRA1-21-1-0014). T.-C.T. gratefully acknowledge the support from The Abdul Latif Jameel Water and Food Systems Laboratory (J-WAFS) Graduate Student Fellowship.
Author information
Authors and Affiliations
Contributions
T.-C.T., E.T., X.L., X.Z. and T.K.L. conceived and designed the research. T.-C.T., E.T., X.L. and H.Y. performed encapsulation and mechanical testing experiments. T.-C.T. and E.T. performed genetic circuit experiments. T.-C.T., E.T. and A.J.R. performed GRO experiments. T.-C.T. and E.T. performed river water experiments. T.-C.T., E.T., X.L., K.Y., A.J.R., C.F.-N., F.J.I., X.Z. and T.K.L. analyzed the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
T.-C.T., E.T., X.L., H.Y., X.Z. and T.K.L. have filed a patent application based on the hydrogel encapsulation technologies with the US Patent and Trademark Office. T.K.L. is a cofounder of Senti Biosciences, Synlogic, Engine Biosciences, Tango Therapeutics, Corvium, BiomX, Eligo Biosciences, Bota.Bio, Avendesora and NE47Bio. T.K.L. also holds financial interests in nest.bio, Armata, IndieBio, MedicusTek, Quark Biosciences, Personal Genomics, Thryve, Lexent Bio, MitoLab, Vulcan, Serotiny, Avendesora and Pulmobiotics.
Additional information
Peer review information Nature Chemical Biology thanks Keekyoung Kim, Minglin Ma and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Responses of encapsulated bacterial cells to external stimuli.
(a) Left: Schematic of GFP expression under the control of an aTc-inducible promoter. Center: Flow cytometry analysis of GFP expression in liquid culture and in hydrogel beads. Samples prepared in triplicate, data represent the mean ±1 s.d. based on analyses of 30000 events. The percentage data were calculated by dividing the numbers of GFP ON cells with the total cell counts. The fold-change data were derived from the mean of fluorescence. Right: Confocal microscopy images of beads encapsulating the aTc-sensing E. coli strain with and without 200 ng/mL aTc. (b) Left: A heme sensing strain which sense heme and generate bioluminescence as an output. The heme released from blood is transported into the cell by ChuA. Middle: Cells retrieved from beads showed a significant increase in luciferase activity. Right: The resulting bioluminescence can be detected with high sensitivity from intact beads. Samples prepared in triplicate, data represent the mean ±1 s.d.
Supplementary information
Supplementary Information
Supplementary Figs. 1–18 and Tables 1 and 2.
Source data
Source Data Fig. 3
Statistical source data and loading curves.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Rights and permissions
About this article
Cite this article
Tang, TC., Tham, E., Liu, X. et al. Hydrogel-based biocontainment of bacteria for continuous sensing and computation. Nat Chem Biol 17, 724–731 (2021). https://doi.org/10.1038/s41589-021-00779-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00779-6
This article is cited by
-
Cas9-assisted biological containment of a genetically engineered human commensal bacterium and genetic elements
Nature Communications (2024)
-
Synthetic microbiology in sustainability applications
Nature Reviews Microbiology (2024)
-
Drinkable in situ-forming tough hydrogels for gastrointestinal therapeutics
Nature Materials (2024)
-
Bioprinting microporous functional living materials from protein-based core-shell microgels
Nature Communications (2023)
-
Machining water through laser cutting of nanoparticle-encased water pancakes
Nature Communications (2023)