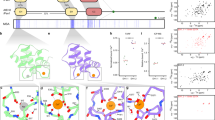
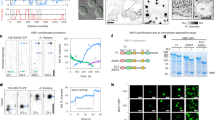
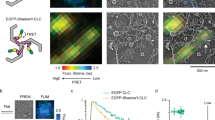
Abstract
Clathrin-mediated endocytosis (CME) is a highly conserved and essential cellular process in eukaryotic cells, but its dynamic and vital nature makes it challenging to study using classical genetics tools. In contrast, although small molecules can acutely and reversibly perturb CME, the few chemical CME inhibitors that have been applied to plants are either ineffective or show undesirable side effects. Here, we identify the previously described endosidin9 (ES9) as an inhibitor of clathrin heavy chain (CHC) function in both Arabidopsis and human cells through affinity-based target isolation, in vitro binding studies and X-ray crystallography. Moreover, we present a chemically improved ES9 analog, ES9-17, which lacks the undesirable side effects of ES9 while retaining the ability to target CHC. ES9 and ES9-17 have expanded the chemical toolbox used to probe CHC function, and present chemical scaffolds for further design of more specific and potent CHC inhibitors across different systems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All accession codes supporting the findings of this study are available within the paper and its Supplementary Information. Structures are accessible under the PDB code 6E4L. There is no restriction on data availability.
References
Reynolds, G. D., Wang, C., Pan, J. & Bednarek, S. Y. Inroads into internalization: five years of endocytic exploration. Plant Physiol. 176, 208–218 (2018).
Kaksonen, M. & Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 19, 313–326 (2018).
Mettlen, M., Chen, P.-H., Srinivasan, S., Danuser, G. & Schmid, S. L. Regulation of clathrin-mediated endocytosis. Annu. Rev. Biochem. 87, 871–896 (2018).
Mishev, K., Dejonghe, W. & Russinova, E. Small molecules for dissecting endomembrane trafficking: a cross-systems view. Chem. Biol. 20, 475–486 (2013).
von Kleist, L. et al. Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell 146, 471–484 (2011).
Macia, E. et al. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10, 839–850 (2006).
McCluskey, A. et al. Building a better dynasore: the Dyngo compounds potently inhibit dynamin and endocytosis. Traffic 14, 1272–1289 (2013).
Elkin, S. R. et al. Ikarugamycin: a natural product inhibitor of clathrin-mediated endocytosis. Traffic 17, 1139–1149 (2016).
Dejonghe, W. et al. Mitochondrial uncouplers inhibit clathrin-mediated endocytosis largely through cytoplasmic acidification. Nat. Commun. 7, 11710 (2016).
Jafari, R. et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 9, 2100 (2014).
Lomenick, B. et al. Target identification using drug affinity responsive target stability (DARTS). Proc. Natl Acad. Sci. USA 106, 21984–21989 (2009).
Jelínková, A. et al. Probing plant membranes with FM dyes: tracking, dragging or blocking? Plant J. 61, 883–892 (2010).
Ludwig, A., Stolz, J. & Sauer, N. Plant sucrose‐H+ symporters mediate the transport of vitamin H. Plant J. 24, 503–509 (2000).
Van Leene, J. et al. An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat. Protoc. 10, 169–187 (2015).
Mishev, K. et al. Nonselective chemical inhibition of Sec7 domain-containing ARF GTPase exchange factors. Plant Cell 30, 2573–2593 (2018).
Kitakura, S. et al. Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23, 1920–1931 (2011).
Popova, N. V., Deyev, I. E. & Petrenko, A. G. Clathrin-mediated endocytosis and adaptor proteins. Acta Naturae 5, 62–73 (2013).
Onizuka, S., Ikewaki, N. & Shiraishi, S. A mechanism by which propofol induces cytotoxicity. J. Drug Metab. Toxicol. 8, 230 (2017).
Poot, M. et al. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J. Histochem. Cytochem. 44, 1363–1372 (1996).
Dolman, N. J., Kilgore, J. A. & Davidson, M. W. A review of reagents for fluorescence microscopy of cellular compartments and structures, part I: BacMam labeling and reagents for vesicular structures. Curr. Protoc. Cytom. 65, 12.30.1–12.30.27 (2013).
Wang, Y.-S., Yoo, C.-M. & Blancaflor, E. B. Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C‐ and N‐termini of the fimbrin actin‐binding domain 2. New Phytol. 177, 525–536 (2008).
Marc, J. et al. GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10, 1927–1940 (1998).
Teh, O.-k & Moore, I. An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448, 493–496 (2007).
Dettmer, J., Hong-Hermesdorf, A., Stierhof, Y.-D. & Schumacher, K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18, 715–730 (2006).
Irani, N. G. et al. Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 8, 583–589 (2012).
Friedrichsen, D. M., Joazeiro, C. A. P., Li, J., Hunter, T. & Chory, J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123, 1247–1255 (2000).
Ortiz-Morea, F. A. et al. Danger-associated peptide signaling in Arabidopsis requires clathrin. Proc. Natl Acad. Sci. USA 113, 11028–11033 (2016).
Gadeyne, A. et al. The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156, 691–704 (2014).
Di Rubbo, S. et al. The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell 25, 2986–2997 (2013).
Dhonukshe, P. et al. Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17, 520–527 (2007).
Adamowski, M. et al. A functional study of AUXILIN-LIKE1 and 2, two putative clathrin uncoating factors in Arabidopsis. Plant Cell 30, 700–716 (2018).
Robinson, D. G. & Pimpl, P. Clathrin and post-Golgi trafficking: a very complicated issue. Trends Plant Sci. 19, 134–139 (2014).
Hicks, G. R. & Raikhel, N. V. Small molecules present large opportunities in plant biology. Annu. Rev. Plant Biol. 63, 261–282 (2012).
Drakakaki, G. et al. Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc. Natl Acad. Sci. USA 108, 17850–17855 (2011).
Zhang, C. et al. Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc. Natl Acad. Sci. USA 113, E41–E50 (2016).
Kania, U. et al. The inhibitor Endosidin 4 targets SEC7 domain-type ARF GTPase exchange factors and interferes with subcellular trafficking in eukaryotes. Plant Cell 30, 2553–2572 (2018).
Li, R. et al. Different endomembrane trafficking pathways establish apical and basal polarities. Plant Cell 29, 90–108 (2017).
Sharfman, M. et al. Endosomal signaling of the tomato leucine-rich repeat receptor-like protein LeEix2. Plant J. 68, 413–423 (2011).
Helsens, K. et al. ms_lims, a simple yet powerful open source laboratory information management system for MS-driven proteomics. Proteomics 10, 1261–1264 (2010).
Scott, W. R. P. et al. The GROMOS biomolecular simulation program package. J. Phys. Chem. 103, 3596–3607 (1999).
Guex, N. & Peitsch, M. C. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997).
Hanwell, M. D. et al. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 4, 17 (2012).
Halgren, T. A. & MMFF, V. I. MMFF94s option for energy minimization studies. J. Comput. Chem. 20, 720–729 (1999).
Sanner, M. F. Python: a programming language for software integration and development. J. Mol. Graph. Model. 17, 57–61 (1999).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Schüttelkopf, A. W. & van Aalten, D. M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D 60, 1355–1363 (2004).
Huynh, K. & Partch, C. L. Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 79, 28.9.1-28.9.14 (2015).
Acknowledgements
We thank S. Vanneste for fruitful discussions, R. Kumar for providing the pDONR211-AP2S plasmid, D. Martinez Molina for help with the CETSA protocol and M. De Cock for help in preparing the manuscript. This work was supported by the Research Foundation-Flanders (project No. G022516N to E.R., project No. G009415N to D.V.D and project No. G0E5718N to E.R. and J.F.); the European Research Council (ERC Co T-Rex, grant No. 682436 to D.V.D); the Deutsche Forschungsgemeinschaft (No. TRR186/A08 to V.H.); the Agency for Innovation by Science and Technology for postdoctoral (K.M.) and predoctoral (W.D. and S.D.M) fellowships; the China Science Council for a predoctoral fellowship (Q.L.); the joint research projects (Nos. VS.025.13N and VS.095.16N) within the framework of cooperation between the Research Foundation-Flanders and the Bulgarian Academy of Sciences (K.M.); and the Belgian Science Policy Office for a postdoctoral fellowship to non-EU researchers (I.S.).
Author information
Authors and Affiliations
Contributions
W.D. and E.R. initiated the work. W.D., I.S. and E.R. designed the experiments. W.D., I.S., B.D., A.M. and J.W. performed SAR. W.D., K.M., A.S. and K.G. performed affinity purification and MS analysis. H.B. and V.H. performed the X-ray crystallography. I.S., S.D.M. and S.N.S. performed the in vitro binding assay. W.N. did the molecular docking. W.D., I.S. and Q.L. performed CETSA. W.D., I.S. and Q.L. performed DARTS. W.D., I.S., E.M., D.V.S., and D.V.D. carried out the imaging and data analysis. I.S. performed the cloning and generated transgenic Arabidopsis cell cultures. W.D., I.S., A.D. and D.A. performed ATP measurements. M.V. and J.F. contributed to the HeLa cell assays. K.Y. generated the TPLATE antibody. Q.L. and R.D.R performed TEM. W.D., I.S. and E.R. wrote the manuscript. All authors commented on the results and the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–12, Supplementary Tables 1–3
Supplementary Note 1
Synthetic Procedures
Rights and permissions
About this article
Cite this article
Dejonghe, W., Sharma, I., Denoo, B. et al. Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat Chem Biol 15, 641–649 (2019). https://doi.org/10.1038/s41589-019-0262-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0262-1
This article is cited by
-
Phytoalexin sakuranetin attenuates endocytosis and enhances resistance to rice blast
Nature Communications (2024)
-
Biomolecular condensation orchestrates clathrin-mediated endocytosis in plants
Nature Cell Biology (2024)
-
Adaptor protein complex interaction map in Arabidopsis identifies P34 as a common stability regulator
Nature Plants (2023)
-
Fungal small RNAs ride in extracellular vesicles to enter plant cells through clathrin-mediated endocytosis
Nature Communications (2023)
-
The endocytic TPLATE complex internalizes ubiquitinated plasma membrane cargo
Nature Plants (2022)