Abstract
The origin and functionality of long noncoding RNA (lncRNA) remain poorly understood. Here, we show that multiple quantitative trait loci modulating distinct domestication traits in soybeans are pleiotropic effects of a locus composed of two tandem lncRNA genes. These lncRNA genes, each containing two inverted repeats, originating from coding sequences of the MYB genes, function in wild soybeans by generating clusters of small RNA (sRNA) species that inhibit the expression of their MYB gene relatives through post-transcriptional regulation. By contrast, the expression of lncRNA genes in cultivated soybeans is severely repressed, and, consequently, the corresponding MYB genes are highly expressed, shaping multiple distinct domestication traits as well as leafhopper resistance. The inverted repeats were formed before the divergence of the Glycine genus from the Phaseolus–Vigna lineage and exhibit strong structure–function constraints. This study exemplifies a type of target for selection during plant domestication and identifies mechanisms of lncRNA formation and action.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data are available in the main text, the Supplementary Information, public databases or referenced studies. All raw sequence data generated in this study have been deposited in the NCBI database under BioProject PRJNA876203. Genotypic data from the USDA soybean germplasm collection used for the GWAS on pubescence form and leafhopper resistance in Extended Data Fig. 1c,d were downloaded from the SoyBase database (https://soybase.org/snps/download.php). Genotypic data of the resequenced soybean accessions used for the GWAS on pubescence form in Extended Data Fig. 1a,b were downloaded from the Genome Variation Map database in BIG Data Center (http://bigd.big.ac.cn/gvm/getProjectDetail?project=GVM000063). RNA-seq, sRNA and WGBS data of the 45 highly diverse soybean accessions were download from the Sequence Read Archive database at NCBI under accession number PRJNA432760 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA432760). Source data are provided with this paper.
Code availability
All software used in this study is publicly available as described in the Methods and the Reporting summary. Detailed parameters used for analyzing each type of sequencing data have been described in the Methods. An in-house Perl scrip used for creating SNP-corrected genomes is available at Zenodo (https://doi.org/10.5281/zenodo.10801184)51.
References
Olsen, K. M. & Wendel, J. F. A bountiful harvest: genomic insights into crop domestication phenotypes. Annu. Rev. Plant Biol. 64, 47–70 (2013).
Doebley, J. F., Gaut, B. S. & Smith, B. D. The molecular genetics of crop domestication. Cell 127, 1309–1321 (2006).
Sedivy, E. J., Wu, F. & Hanzawa, Y. Soybean domestication: the origin, genetic architecture and molecular bases. New Phytol. 214, 539–553 (2017).
Swarm, S. A. et al. Genetic dissection of domestication-related traits in soybean through genotyping-by-sequencing of two interspecific mapping populations. Theor. Appl. Genet. 132, 1195–1209 (2019).
Broersma, D., Bernard, R. & Luckmann, W. Some effects of soybean pubescence on populations of the potato leafhopper. J. Econ. Entomol. 65, 78–82 (1972).
Liu, Y. et al. Pan-genome of wild and cultivated soybeans. Cell 182, 162–176 (2020).
Song, Q. et al. Fingerprinting soybean germplasm and its utility in genomic research. G3 5, 1999–2006 (2015).
Shen, Y. et al. DNA methylation footprints during soybean domestication and improvement. Genome Biol. 19, 128 (2018).
Schmutz, J. et al. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 (2010).
Choi, H.-K. et al. Estimating genome conservation between crop and model legume species. Proc. Natl Acad. Sci. USA 101, 15289–15294 (2004).
Zheng, F. et al. Molecular phylogeny and dynamic evolution of disease resistance genes in the legume family. BMC Genomics 17, 402 (2016).
Vaucheret, H. & Fagard, M. Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 17, 29–35 (2001).
Statello, L., Guo, C.-J., Chen, L.-L. & Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 (2021).
Parniske, M. et al. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91, 821–832 (1997).
Reams, A. B. & Roth, J. R. Mechanisms of gene duplication and amplification. Cold Spring Harb. Perspect. Biol. 7, a016592 (2015).
Cuerda-Gil, D. & Slotkin, R. K. Non-canonical RNA-directed DNA methylation. Nat. Plants 2, 16163 (2016).
Gagliardi, D. et al. Dynamic regulation of chromatin topology and transcription by inverted repeat-derived small RNAs in sunflower. Proc. Natl Acad. Sci. USA 116, 17578–17583 (2019).
Lu, C. et al. Miniature inverted-repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol. Biol. Evol. 29, 1005–1017 (2012).
Arce, A. L. et al. Polymorphic inverted repeats near coding genes impact chromatin topology and phenotypic traits in Arabidopsis thaliana. Cell Rep. 42, 112029 (2023).
Wu, N. et al. A MITE variation‐associated heat‐inducible isoform of a heat‐shock factor confers heat tolerance through regulation of JASMONATE ZIM‐DOMAIN genes in rice. New Phytol. 234, 1315–1331 (2022).
Niu, C. et al. Methylation of a MITE insertion in the MdRFNR1-1 promoter is positively associated with its allelic expression in apple in response to drought stress. Plant Cell 34, 3983–4006 (2022).
Xu, L. et al. Regulation of rice tillering by RNA-directed DNA methylation at miniature inverted-repeat transposable elements. Mol. Plant 13, 851–863 (2020).
Bradley, D. et al. Evolution of flower color pattern through selection on regulatory small RNAs. Science 358, 925–928 (2017).
Fabian, M. R. & Sonenberg, N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 19, 586–593 (2012).
Doebley, J., Stec, A. & Hubbard, L. The evolution of apical dominance in maize. Nature 386, 485–488 (1997).
Tan, L. et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40, 1360–1364 (2008).
R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing (2013).
Zeng, Z.-B. Precision mapping of quantitative trait loci. Genetics 136, 1457–1468 (1994).
Broman, K. W., Wu, H., Sen, Ś. & Churchill, G. A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890 (2003).
Bradbury, P. J. et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635 (2007).
Yu, J. et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208 (2006).
Zhou, Z. et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 33, 408–414 (2015).
Lei, Y. et al. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7, 1494–1496 (2014).
Bai, M. et al. Generation of a multiplex mutagenesis population via pooled CRISPR–Cas9 in soya bean. Plant Biotechnol. J. 18, 721–731 (2020).
Richter, G. L. et al. Estimating leaf area of modern soybean cultivars by a non-destructive method. Bragantia 73, 416–425 (2014).
Abràmoff, M. D., Magalhães, P. J. & Ram, S. J. Image processing with ImageJ. Biophotonics Int. 11, 36–42 (2004).
Chen, C. et al. Real-time quantification of microRNAs by stem-loop RT–PCR. Nucleic Acids Res. 33, e179 (2005).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Addo-Quaye, C., Miller, W. & Axtell, M. J. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25, 130–131 (2009).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27, 1571–1572 (2011).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Zhang, Y. et al. Model-based analysis of ChIP–seq (MACS). Genome Biol. 9, R137 (2008).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Maere, S., Heymans, K. & Kuiper, M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 (2005).
Tamura, K. & Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526 (1993).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Dai, X., Zhuang, Z. & Zhao, P. X. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46, W49–W54 (2018).
Wang, X. An in-house Perl script used for creating SNP-corrected references. Zenodo https://doi.org/10.5281/zenodo.10801184 (2024).
Acknowledgements
We thank X. Chen, D. Lisch and R. Schmitz for constructive comments on this work. This work was mainly supported by the Agriculture and Food Research Initiative of the USDA National Institute of Food and Agriculture (grants 2018-67013-27425, 2021-67013-33722 and 2022-67013-37037) and partially supported by the United Soybean Board, the North Central Soybean Research Program, the Indiana Soybean Alliance and Ag Alumni Seed.
Author information
Authors and Affiliations
Contributions
J.M. and Xianzhong Feng designed the research. W.W., J.D., Xingxing Feng, X.W., L.C., C.B.C., S.A.S., R.L.N., S.L. and J.W. performed experiments. W.W., X.W., B.C.M. and J.M. analyzed data. W.W. and J.M. wrote the manuscript, and B.C.M. edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Yong-Qiang An and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Association studies, selection analyses and expression analyses.
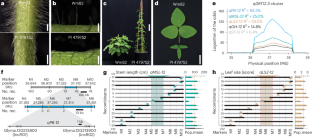
a-b, Genome-wide association study (GWAS) on pubescence form using the re-sequencing data from 74 G. soja and 594 G. max accessions6 and corresponding phenotypic data from the USDA soybean germplasm database (Supplementary Table 2). The red color highlights markers within the fine-mapped qDRT12.3 region. c-d, GWAS on leafhopper resistance (c) and pubescence form (d) using the genotypic data from 784 soybean accession7 and corresponding phenotypic data from the USDA database (Supplementary Table 4). The rectangle highlights the qDRT12.3 locus. In (a-d), the P values were determined by the F-test for each marker. e, Frequencies of erect and appressed pubescence form in G. soja, landrace and elite cultivar sub-populations6. n indicates the number of soybean accessions in each sub-population. f, Selective sweep surrounding the qDRT12.3 region. The y-axis is the ratio of nucleotide diversity (π) of landraces (n = 328) with erect pubescence over G. soja (n = 103)6 calculated for every 100-kb window with 10-kb sliding steps. Each vertical bar represents the value at the middle point of each sliding window. The red arrows pinpoint the positions of lncRG1 and lncRG2. The x-axis presents the physical positions based on the Zhonghuang 13 (v2) genome assembly. g, Expression levels of lncRG1 and lncRG2 in the V1-stage stem tips of G. soja (n = 9) and G. max (n = 36) (Supplementary Table 7). The expression levels were measured with RNA-seq data8 and represented as mean ± SEM. FPKM, fragments per kilobase of transcript per million mapped reads. The dots indicate the values from biologically independent samples (n = 3). The numbers above the bars are P values determined by a two-sided Student’s t-test. h, Co-expression between lncRG1 and lncRG2 in the V1-stage stem tips. The expression levels of lncRG1 and lncRG2 were measured with the RNA-seq data8. Each dot represents a single soybean accession, with blue dots for G. soja haplotype (n = 11) and orange dots for G. max haplotype (n = 34). Dashed line is the trend line. The P value is obtained by a two-sided Pearson’s correlation test. i, Collinearity between the lncRG1-lncRG2 region and the lncRG3-lncRG4 region. Boxes represent genes and grey shades connect WGD pairs.
Extended Data Fig. 2 Abundance and distribution of sRNAs produced by lncRG1 and lncRG2 in a pair of RILs and the transgenic lines, and images of transgenic lines.
a, Abundance and distribution of sRNAs produced by lncRG1 in RIL186 (qdrt12.3) and RIL334 (qDRT12.3). The x-axis shows the position on the lncRG1 transcript, and the y-axis is the abundance in copy per million reads (CPM). b, Abundance and distribution of sRNAs produced by lncRG2 in RIL186 (qdrt12.3) and RIL334 (qDRT12.3). The x-axis shows the position on the lncRG2 transcript, and the y-axis is abundance in copy per million reads (CPM). c, Frequencies of sRNA from lncRG1 at different sizes from 17nt to 25nt in RIL186 (qdrt12.3) and RIL334 (qDRT12.3). d, Frequencies of sRNA from lncRG2 at different sizes 17nt to 25nt in RIL186 (qdrt12.3) and RIL334 (qDRT12.3). e, Abundance and distribution of sRNAs along the transcript of lncRG1 in the lncRG1-LOOPOE transgenic lines. The x-axis shows the position on the lncRG1 transcript, and the y-axis is the abundance in copy per million reads (CPM). f, Abundance and distribution of sRNAs along the transcript of lncRG2 in the lncRG2-LOOPOE transgenic lines. The x-axis shows the position on the lncRG2 transcript, and the y-axis is the abundance in copy per million reads (CPM). g, Plant images of the transgenic lines that overexpress the inverted repeats of lncRG1 and lncRG2. Bars = 10 cm. h, Leaf images of the transgenic lines that overexpress the inverted repeats of lncRG1 and lncRG2. Bars=5 cm. i, Relative expression levels of the predicted CDS of lncRG1 and lncRG2 in the transgenic lines that overexpress the predicted CDS, as determined by qRT-PCR with Wm82 set as “1” and the others adjusted accordingly. The dots show the values from biologically independent samples (n = 3). Data are represented as mean ± SEM. j, images of the transgenic plants that overexpress the predicted CDS of lncRG1 and lncRG2, Bars = 5 mm, 5 cm, 5 cm in top, middle and bottom, respectively.
Extended Data Fig. 3 Mutations created by CRISPR-Cas9, protein-protein interaction as detected by Y2H and ChIP-seq analysis.
a-c, Frameshift mutants created by CRISPR-Cas9 for each of the three MYB genes, Glyma.01G051700 (a), Glyma.02G110000 (b) and Glyma.02G110100 (c). The top sequence shows the Wm82 sequence and the position of each base pair in Wm82. - represent deletions in the editing lines. Red asterisk indicates the lines selected for crossing to make double editing lines. d, Primary Y2H tests to confirm whether the MYB target genes can active the reporter gene. EV represents empty vector. e, Protein-protein interactions among MYB transcription factors as detected by the yeast two hybrid (Y2H) system. Colonies on DDO plate indicate the successful transformation of the construct in yeast cells. Blue colonies on QDO/X/A plates indicate positive protein-protein interactions. AD, activation domain; BD, binding domain; DDO, double dropout; QDO, quadruple dropout. X, X-alpha-Gal; A, Aureobasidin A. f-g, Distribution of the locations of the ChIP-seq peaks relative to target genes detected in the Glyma.01G051700-FLAG and Glyma.02G110000-FLAG transgenic lines, respectively. h-i, Frequency of the ChIP-Seq peaks surrounding the transcription start sites (SST) detected in the Glyma.01G051700-FLAG and Glyma.02G110000-FLAG transgenic lines, respectively. j, Number of potential downstream genes identified by ChIP-seq in the Glyma.01G051700-FLAG and Glyma.02G110000-FLAG transgenic lines. k, Gene ontology (GO) classification for the genes detected in both the Glyma.01G051700-FLAG and Glyma.02G110000-FLAG transgenic lines. The P value was determined by Fisher’s exact test adjusted for false discovery rate.
Extended Data Fig. 4 Copy number conservation of lncRG1 and lncRG2 in the soybean pan-genome and evolution of lncRG3 and lncRG4.
a, Genomic sequence and gene alignments among the soybean pan-genome accessions at the lncRG1-lncRG2 region, including flanking genes. Boxes represent genes and grey color indicate syntenic blocks among genomes. b, Relative expression levels of lncRG1, lncRG2, lncRG3 and lncRG4 in the stem tips of Wm82 and PI 479752, as determined by qRT-PCR. The dots show the values from biologically independent samples (n = 3). Data are represented as mean ± SEM. c-d, Secondary structures of lncRG3 and lncRG4 and the sRNAs mapped to their inverted repeats. e, nucleotide diversity within the inverted repeats of lncRG1, lncRG2, lncRG3 and lncRG4. The dots show the values of nucleotide diversity calculated from different soybean pan-genome accessions (n = 27). The horizontal lines indicate the medians, and the boxes represent the interquartile range (IQR). The whiskers represent the range of 1.5 times IQR and dots beyond the whiskers are outlier values. The numbers above the boxes are P values determined by a two-sided Student’s t-test.
Extended Data Fig. 5 Distribution of the sRNAs produced by lncRG1 and lncRG2 in ten diverse soybean accessions.
The x-axis shows the position on the lncRG1 (a) or lncRG2 (b) transcripts, and the y-axis is abundance in copy per million reads (CPM). The relative abundances of sRNAs of different sizes detected in individual accessions (Supplementary Table 7) are shown in percentage (%) in individual pies.
Extended Data Fig. 6 Association between epigenetic variations and expression levels of lncRG1 and lncRG2.
a, Differences of CpG, CHG and CHH DNA methylation between the G. max haplotype (n = 29) and the G. soja haplotype (n = 10) surrounding lncRG1 and lncRG2 (Supplementary Table 7). Each vertical bar represents the average methylation level difference within a 300 bp window between the two haplotypes with sliding step=50 bp. The purple color highlights the differences in the promoter regions of the two genes. The red asterisk indicates the window used for correlation analysis in (b) and (c). b-c, Correlations between the CpG methylation differences in the promoter regions of lncRG1 and lncRG2 with their expression levels as measured by Pearson’s correlation coefficient (n = 41). The P values are obtained by a two-sided Pearson’s correlation test. Dashed lines are the trend lines.
Supplementary information
Supplementary Tables
Supplementary Table 1. List of recombinants, including genotypic and phenotypic data, used for fine mapping. Supplementary Table 2. List of resequenced lines used for association mapping on pubescence form. Supplementary Table 3. Results from the GWAS on pubescence form using the resequenced lines (the P value was calculated from an F-test for each marker). Supplementary Table 4. List and phenotypic values of the USDA soybean accessions used for GWAS on leafhopper resistance and pubescence form. Supplementary Table 5. Results from the GWAS on leafhopper resistance and pubescence form using USDA soybean accessions (the P value was calculated from an F-test for each marker). Supplementary Table 6. Expression levels (FPKM) of lncRG genes in shoots, stems and leaves of Wm82 and PI 479752. Supplementary Table 7. List of 45 diverse soybean accessions with RNA-seq, sRNA and bisulfite-seq data available from a previous study. Supplementary Table 8. sRNA species produced by lncRG1 and lncRG2 with CPM > 10. Supplementary Table 9. List of genes targeted by 27 sRNA species (CPM > 100) produced by lncRG1 and lincRG2. Supplementary Table 10. Expression levels (FPKM) of the 163 target genes in shoots, stems and leaves of Wm82 and PI 479752. Supplementary Table 11. List of peaks detected by ChIP–seq in Glyma.01G051700-FLAG-transgenic lines (the P value was calculated from a Poisson test for each region). Supplementary Table 12. List of peaks detected by ChIP–seq in Glyma.02G110000-FLAG-transgenic lines (the P value was calculated from a Poisson test for each region). Supplementary Table 13. List of top 20 sRNA species produced by lncRG1 and lncRG2 in ten diverse soybean accessions with the G. soja haplotype. Supplementary Table 14. List of genes targeted by sRNA species (top 20) produced by lncRG1 and lincRG2 in ten soybean accessions. Supplementary Table 15. List of primers used in this study.
Supplementary Video 1
Supplementary Video 1. Appressed pubescence in a double mutant attributed to susceptibility to leafhopper.
Supplementary Video 2
Supplementary Video 2. Erected pubescence in Wm82 attributed to resistance to leafhopper.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed gel image.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, W., Duan, J., Wang, X. et al. Long noncoding RNAs underlie multiple domestication traits and leafhopper resistance in soybean. Nat Genet (2024). https://doi.org/10.1038/s41588-024-01738-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41588-024-01738-2



