Abstract
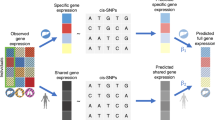
Translating genome-wide association study (GWAS) loci into causal variants and genes requires accurate cell-type-specific enhancer–gene maps from disease-relevant tissues. Building enhancer–gene maps is essential but challenging with current experimental methods in primary human tissues. Here we developed a nonparametric statistical method, SCENT (single-cell enhancer target gene mapping), that models association between enhancer chromatin accessibility and gene expression in single-cell or nucleus multimodal RNA sequencing and ATAC sequencing data. We applied SCENT to 9 multimodal datasets including >120,000 single cells or nuclei and created 23 cell-type-specific enhancer–gene maps. These maps were highly enriched for causal variants in expression quantitative loci and GWAS for 1,143 diseases and traits. We identified likely causal genes for both common and rare diseases and linked somatic mutation hotspots to target genes. We demonstrate that application of SCENT to multimodal data from disease-relevant human tissue enables the scalable construction of accurate cell-type-specific enhancer–gene maps, essential for defining noncoding variant function.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The publicly available datasets were downloaded via Gene Expression Omnibus (accession codes GSE140203, GSE156478, GSE178707, GSE194122, GSE193240 and GSE178453) or web repository (https://www.10xgenomics.com/resources/datasets?query=&page=1&configure%5Bfacets%5D%5B0%5D=chemistryVersionAndThroughput&configure%5Bfacets%5D%5B1%5D=pipeline.version&configure%5BhitsPerPage%5D=500&menu%5Bproducts.name%5D=Single%20Cell%20Multiome%20ATAC%20%2B%20Gene%20Expression). The raw data for arthritis-tissue dataset (single-cell multimodal RNA/ATAC–seq and single-cell ATAC–seq) are deposited at the NIH Database of Genotypes and Phenotypes (dbGaP accession number phs003417.v1.p1) and the Gene Expression Omnibus (GEO accession number GSE243917).
Code availability
The computational scripts related to this manuscript are available at https://github.com/immunogenomics/SCENT (https://doi.org/10.5281/zenodo.10452116)124.
References
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP–trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
Visscher, P. M. et al. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet 101, 5–22 (2017).
Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019).
Claussnitzer, M. et al. A brief history of human disease genetics. Nature 577, 179–189 (2020).
Plenge, R. M., Scolnick, E. M. & Altshuler, D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 12, 581–594 (2013).
Shendure, J., Findlay, G. M. & Snyder, M. W. Genomic medicine—progress, pitfalls, and promise. Cell 177, 45–57 (2019).
Schaid, D. J., Chen, W. & Larson, N. B. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 19, 491–504 (2018).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Edwards, S. L., Beesley, J., French, J. D. & Dunning, M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet 93, 779–797 (2013).
Trynka, G. et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 45, 124–130 (2013).
Sanyal, A., Lajoie, B. R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012).
Smemo, S. et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375 (2014).
Won, H. et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527 (2016).
Strober, B. J. et al. Dynamic genetic regulation of gene expression during cellular differentiation. Science 364, 1287–1290 (2019).
Cuomo, A. S. E. et al. Single-cell RNA-sequencing of differentiating iPS cells reveals dynamic genetic effects on gene expression. Nat. Commun. 11, 810 (2020).
Zhernakova, D. V. et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 49, 139–145 (2017).
Nathan, A. et al. Single-cell eQTL models reveal dynamic T cell state dependence of disease loci. Nature 606, 120–128 (2022).
Wakefield, J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am. J. Hum. Genet. 81, 208–227 (2007).
Maller, J. B. et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 44, 1294–1301 (2012).
Hormozdiari, F., Kostem, E., Kang, E. Y., Pasaniuc, B. & Eskin, E. Identifying causal variants at loci with multiple signals of association. Genetics 198, 497–508 (2014).
Benner, C. et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501 (2016).
Wang, G., Sarkar, A., Carbonetto, P. & Stephens, M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J. R. Stat. Soc. Ser. B 82, 1273–1300 (2020).
Weissbrod, O. et al. Functionally informed fine-mapping and polygenic localization of complex trait heritability. Nat. Genet. 52, 1355–1363 (2020).
Wojcik, G. L. et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 570, 514–518 (2019).
Chen, M. H. et al. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell 182, 1198–1213.e14 (2020).
Ishigaki, K. et al. Multi-ancestry genome-wide association analyses identify novel genetic mechanisms in rheumatoid arthritis. Nat. Genet. 54, 1640–1651 (2022).
Kichaev, G. & Pasaniuc, B. Leveraging functional-annotation data in trans-ethnic fine-mapping studies. Am. J. Hum. Genet. 97, 260–271 (2015).
Kanai, M. et al. Insights from complex trait fine-mapping across diverse populations. Preprint at medRxiv https://doi.org/10.1101/2021.09.03.21262975 (2021).
Huang, H. et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178 (2017).
Farh, K. K. H. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2014).
Mahajan, A. et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 50, 1505–1513 (2018).
Kichaev, G. et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 10, e1004722 (2014).
Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Chen, L. et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell 167, 1398–1414.e24 (2016).
Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Boix, C. A., James, B. T., Park, Y. P., Meuleman, W. & Kellis, M. Regulatory genomic circuitry of human disease loci by integrative epigenomics. Nature 590, 300–307 (2021).
Fulco, C. P. et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664 (2019).
Nasser, J. et al. Genome-wide enhancer maps link risk variants to disease genes. Nature 593, 238–243 (2021).
Gazal, S. et al. Combining SNP-to-gene linking strategies to identify disease genes and assess disease omnigenicity. Nat. Genet. 54, 827–836 (2022).
Pickar-Oliver, A. & Gersbach, C. A. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 20, 490–507 (2019).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Baglaenko, Y., Macfarlane, D., Marson, A., Nigrovic, P. A. & Raychaudhuri, S. Genome editing to define the function of risk loci and variants in rheumatic disease. Nat. Rev. Rheumatol. 17, 462–474 (2021).
Cao, J. et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380–1385 (2018).
Chen, S., Lake, B. B. & Zhang, K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 37, 1452–1457 (2019).
Ma, S. et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 183, 1103–1116.e20 (2020).
Allaway, K. C. et al. Genetic and epigenetic coordination of cortical interneuron development. Nature 597, 693–697 (2021).
Trevino, A. E. et al. Chromatin and gene-regulatory dynamics of the developing human cerebral cortex at single-cell resolution. Cell 184, 5053–5069.e23 (2021).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021).
Pliner, H. A. et al. Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol. Cell 71, 858–871.e8 (2018).
Lähnemann, D. et al. Eleven grand challenges in single-cell data science. Genome Biol. 21, 31 (2020).
Sarkar, A. & Stephens, M. Separating measurement and expression models clarifies confusion in single-cell RNA sequencing analysis. Nat. Genet. 53, 770–777 (2021).
Chen, H. et al. Assessment of computational methods for the analysis of single-cell ATAC–seq data. Genome Biol. 20, 1–25 (2019).
Granja, J. M. et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol. 37, 1458–1465 (2019).
Townes, F. W., Hicks, S. C., Aryee, M. J. & Irizarry, R. A. Feature selection and dimension reduction for single-cell RNA-seq based on a multinomial model. Genome Biol. 20, 1–16 (2019).
Efron, B. & Tibshirani, R. J. An Introduction to the Bootstrap (Chapman and Hall, 1994).
Weinand, K. et al. The chromatin landscape of pathogenic transcriptional cell states in rheumatoid arthritis. Preprint at bioRxiv https://doi.org/10.1101/2023.04.07.536026 (2023).
Luecken, M. D. et al. A sandbox for prediction and integration of DNA, RNA, and proteins in single cells. In 35th Conference on Neural Information Processing Systems Datasets and Benchmarks Track (Round 2) (NeurIPS, 2021).
Mimitou, E. P. et al. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol. 39, 1246–1258 (2021).
Chen, A. F. et al. NEAT-seq: simultaneous profiling of intra-nuclear proteins, chromatin accessibility and gene expression in single cells. Nat. Methods 19, 547–553 (2022).
Meijer, M. et al. Epigenomic priming of immune genes implicates oligodendroglia in multiple sclerosis susceptibility. Neuron 110, 1193–12 (2022).
Zhang, Z. et al. Single nucleus transcriptome and chromatin accessibility of postmortem human pituitaries reveal diverse stem cell regulatory mechanisms. Cell Rep. https://doi.org/10.1016/J.CELREP.2022.110467 (2022).
Abascal, F. et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020).
Westra, H. J. & Franke, L. From genome to function by studying eQTLs. Biochim. Biophys. Acta 1842, 1896–1902 (2014).
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005).
Hujoel, M. L. A., Gazal, S., Hormozdiari, F., van de Geijn, B. & Price, A. L. Disease heritability enrichment of regulatory elements is concentrated in elements with ancient sequence age and conserved function across species. Am. J. Hum. Genet 104, 611–624 (2019).
Mumbach, M. R. et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet. 49, 1602–1612 (2017).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Wang, X. & Goldstein, D. B. Enhancer domains predict gene pathogenicity and inform gene discovery in complex disease. Am. J. Hum. Genet. 106, 215–233 (2020).
Aguet, F. et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Wang, Q. S. et al. Leveraging supervised learning for functionally informed fine-mapping of cis-eQTLs identifies an additional 20,913 putative causal eQTLs. Nat. Commun. 12, 3394 (2021).
Zou, J. et al. Leveraging allelic imbalance to refine fine-mapping for eQTL studies. PLoS Genet. 15, e1008481 (2019).
Chen, W., McDonnell, S. K., Thibodeau, S. N., Tillmans, L. S. & Schaid, D. J. Incorporating functional annotations for fine-mapping causal variants in a Bayesian framework using summary statistics. Genetics 204, 933–958 (2016).
Gaffney, D. J. et al. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 13, R7 (2012).
Göring, H. H. H. et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 39, 1208–1216 (2007).
Wen, X., Luca, F. & Pique-Regi, R. Cross-population joint analysis of eQTLs: fine mapping and functional annotation. PLoS Genet. 11, e1005176 (2015).
Kurki, M. I. et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518 (2023).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Dey, K. K. et al. SNP-to-gene linking strategies reveal contributions of enhancer-related and candidate master-regulator genes to autoimmune disease. Cell Genomics 2, 100145 (2022).
Freund, M. K. et al. Phenotype-specific enrichment of mendelian disorder genes near GWAS regions across 62 complex traits. Am. J. Hum. Genet. 103, 535–552 (2018).
Gate, R. E. et al. Genetic determinants of co-accessible chromatin regions in activated T cells across humans. Nat. Genet. 50, 1140–1150 (2018).
Khetan, S. et al. Type 2 diabetes-associated genetic variants regulate chromatin accessibility in Human Islets. Diabetes 67, 2466–2477 (2018).
Alasoo, K. et al. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet. 50, 424–431 (2018).
Currin, K. W. et al. Genetic effects on liver chromatin accessibility identify disease regulatory variants. Am. J. Hum. Genet. 108, 1169–1189 (2021).
Kumasaka, N., Knights, A. J. & Gaffney, D. J. Fine-mapping cellular QTLs with RASQUAL and ATAC–seq. Nat. Genet. 48, 206–213 (2015).
Kerimov, N. et al. A compendium of uniformly processed human gene expression and splicing quantitative trait loci. Nat. Genet. 53, 1290–1299 (2021).
Sagara, H. et al. Activation of TGF-β/Smad2 signaling is associated with airway remodeling in asthma. J. Allergy Clin. Immunol. 110, 249–254 (2002).
Chiou, J. et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 594, 398–402 (2021).
Mouri, K. et al. Prioritization of autoimmune disease-associated genetic variants that perturb regulatory element activity in T cells. Nat. Genet. 54, 603–612 (2022).
Javierre, B. M. et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–1384.e19 (2016).
Radtke, F., Fasnacht, N. & MacDonald, H. R. Notch signaling in the immune system. Immunity 32, 14–27 (2010).
Wei, K. et al. Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature 582, 259–264 (2020).
Delacher, M. et al. Rbpj expression in regulatory T cells is critical for restraining TH2 responses. Nat. Commun. 10, 1621 (2019).
Blake, J. A. et al. Mouse Genome Database (MGD): knowledgebase for mouse–human comparative biology. Nucleic Acids Res. 49, D981–D987 (2021).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Hillier, S. G. Gonadotropic control of ovarian follicular growth and development. Mol. Cell. Endocrinol. 179, 39–46 (2001).
Rubinstein, W. S. et al. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res. 41, D925–D935 (2013).
Retterer, K. et al. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 18, 696–704 (2016).
Adams, D. R. & Eng, C. M. Next-generation sequencing to diagnose suspected genetic disorders. N. Engl. J. Med. 379, 1353–1362 (2018).
Srivastava, S. et al. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet. Med. 21, 2413–2421 (2019).
Landrum, M. J. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018).
Glocker, E.-O. et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361, 2033–2045 (2009).
Dietlein, F. et al. Genome-wide analysis of somatic noncoding mutation patterns in cancer. Science 376, eabg5601 (2022).
Connally, N. et al. The missing link between genetic association and regulatory function. eLife 11, e74970 (2022).
Dixit, A. et al. Perturb-seq: dissecting molecular circuits with scalable single cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866 (2016).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Morris, J. A. et al. Discovery of target genes and pathways at GWAS loci by pooled single-cell CRISPR screens. Science 380, eadh7699 (2023).
Donlin, L. T. et al. Methods for high-dimensional analysis of cells dissociated from cyropreserved synovial tissue. Arthritis Res Ther. 20, 139 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
PhastCons scores for multiple alignments of 99 vertebrate genomes to the human genome. UCSC Genome Browser https://hgdownload.cse.ucsc.edu/goldenpath/hg19/phastCons100way/ (2014).
gnomAD database. Broad Institute https://gnomad.broadinstitute.org/downloads (2023).
GWAS fine-mapping results. Finucane Lab https://www.finucanelab.org/data (2019).
EpiMap Gene-Enhancer links. Broad Institute https://personal.broadinstitute.org/cboix/epimap/links/pergroup/ (2021).
ABC predictions across 131 biosamples. Broad Institute ftp://ftp.broadinstitute.org/outgoing/lincRNA/ABC/AllPredictions.AvgHiC.ABC0.015.minus150.ForABCPaperV3.txt.gz (2021).
Delaneau, O., Marchini, J. & Zagury, J. F. A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2012).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Gibbs, R. A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
van de Geijn, B., Mcvicker, G., Gilad, Y. & Pritchard, J. K. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods 12, 1061–1063 (2015).
van der Auwera G. & O’Connor, B. Genomics in the Cloud (O’Reilly Media, Inc., 2020).
ClinVar variants. ClinVar https://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh38/clinvar.vcf.gz (2023).
Sakaue, S. immunogenomics/SCENT: v1.0.0. Zenodo https://doi.org/10.5281/zenodo.10452116 (2024).
Acknowledgements
We sincerely thank participants of this study who provided tissue samples. We thank A. Gupta, J. Kang and K. Lagattuta for their comments and helpful discussion on the manuscript. This work is supported in part by funding from the National Institutes of Health (R01AR063759, U01HG012009 and UC2AR081023 to S.R.). S.S. was in part supported by the Uehara Memorial Foundation and The Osamu Hayaishi Memorial Scholarship. K. Weinand was supported by NIH NIAMS T32AR007530. K. Wei was supported by a Burroughs Wellcome Fund Career Awards for Medical Scientists, a Doris Duke Charitable Foundation Clinical Scientist Development Award, a Rheumatology Research Foundation Innovative Research Award, and NIH NIAMS K08AR077037. We thank the Brigham and Women’s Hospital Center for Cellular Profiling Single Cell Multiomics Core for experimental design and protocol optimization. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Consortia
Contributions
S.S. and S.R. conceived the work and wrote the manuscript with critical input from co-authors. S.S. and K. Weinand analyzed the arthritis-tissue dataset and S.S. analyzed publicly available datasets with help and guidance from K.K.D., K.J., M.K., A.M., A.L.P. and S.R. G.F.M.W., Z.Z., M.B.B., L.T.D. and K. Wei provided samples and generated the arthritis-tissue dataset. S.I. refactored the SCENT software implementation as an R package.
Corresponding author
Ethics declarations
Competing interests
S.R. is a founder for Mestag, Inc., a scientific advisor for Rheos, Jannsen and Pfizer, and serves as a consultant for Sanofi and Abbvie. The other authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Tim Stuart and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Distribution of gene expression counts in single-cell RNA-seq and statistics from association between gene expression and chromatin accessibility under null simulation.
a. In an example dataset of arthritis-dataset, mean gene count was strongly correlated with standard deviation of the gene count. b. The correlation between max expression count per gene (x-axis) and the mean naïve association chi-square values (χ2) from Poisson regression between gene expression and chromatin accessibility under null simulation (y-axis). c. The quantile-quantile (QQ) plot of two-sided P values from the Poisson regression between gene expression count and chromatin accessibility under null simulation. d. The QQ plot of two-sided P values from the negative binomial regression between gene expression count and chromatin accessibility under null simulation. e. The QQ plot of two-sided P values from the linear regression between log-normalized and inverse-normal-transformed gene expression and chromatin accessibility under null simulation. f. The QQ plot of two-sided P values estimated from bootstrapping based on the statistics distributions from the Poisson regression between gene expression count and chromatin accessibility under null simulation. g. The QQ plot of two-sided P values estimated from bootstrapping based on the statistics distributions from the negative binomial regression between gene expression count and chromatin accessibility under null simulation. h. Computational runtime benchmarking for Poisson regression with binarized ATAC-seq peak (red), negative binomial regression with binarized ATAC-seq peak (teal), and Poisson regression with non-binarized ATAC-seq peak (blue). The values are relative to the computational time for Poisson regression, and bars are the mean across n=100 randomly selected peak-gene pairs. Horizontal lines (error bars) indicate one standard deviation from the mean.
Extended Data Fig. 2 Schematic overview of SCENT model using Poisson regression and non-parametric bootstrapping.
We first run Poisson regression associating the raw gene expression count (RNA-seq) with the peak accessibility (ATAC-seq) accounting for technical covariates across the entire cells in the multimodal data to estimate βpeak. Then, we resampled cells with replacement from the full data in each of the bootstrapping round and re-estimated \({\beta {\prime} }_{{peak}}\) for N times. We compared this empirical distribution of \({\beta {\prime} }_{{peak}}\) against the null hypothesis (\({\beta {\prime} }_{{peak}}\) = 0) to derive the significance of βpeak (that is, two-sided bootstrapping-based P value = Pbootstrap).
Extended Data Fig. 3 The QQ plot of SCENT P values by bootstrapping.
We applied SCENT to each of 23 broad cell types from 9 single-cell multimodal datasets. Each QQ plot represents two-sided Pbootstrap values in each cell type in each dataset (a. arthritis-tissue, b. public PBMC, c. NeurIPS, d. SHARE-seq, e. Dogma-seq (control), f. Dogma-seq (stimulated) g. NEAT-seq, h. Brain, i. Pituitary.
Extended Data Fig. 4 Properties of SCENT peaks.
a. The number of significant SCENT peaks per gene across genes we investigated in at least one dataset-cell type pair. b. The number of significant gene-peak pairs discovered by SCENT with FDR < 10% in each dataset (y-axis) as a function of the total number of ATAC-seq fragments in each dataset (x-axis), colored by the dataset. c. The number of significant gene-peak pairs discovered by SCENT with FDR < 10% in each dataset (y-axis) as a function of the total number of unique RNA molecules in each dataset (x-axis), colored by the dataset. d. The effect size correlation r by Pearson’s correlation between arthritis-tissue dataset and the other dataset for the same cell type (left) and the directional (sign) concordance between arthritis-tissue dataset and the other dataset for the same cell type (right). e. Fraction of overlap with ENCODE cCREs in SCENT (teal) or non-SCENT peaks (orange) in each dataset and random set of cis-non-coding regions (pink). f. The mean Δ phastCons score for SCENT with excluding promoter peaks (teal) and all cis-ATAC peaks with excluding promoter peaks (yellow) in each of the three example multimodal datasets. The bars indicate the 95% CI by bootstrapping genes (nbootstrap=1000). g. The mean Δ phastCons score between SCENT peaks and TSS-distance-matched non-SCENT peaks across all the genes. The bars indicate the 95% CI by bootstrapping genes (nbootstrap=1000).
Extended Data Fig. 5 Mutational constraint on genes with a high number of SCENT peaks.
For each gene, the number of SCENT peaks were counted and binned as shown in the x-asis, and mutational constraint metric (pLI (the probability of being loss of function intolerant): a, LOEUF (the loss-of-function observed/expected upper bound fraction): b) for genes within each bin are shown as a violin plot on the y-axis. The dots indicate the mean score in each bin, and the error bars indicate one standard deviation from the mean. Each bin consists of 555-4071 genes in a and 568-4265 genes in b.
Extended Data Fig. 6 Causal variant enrichment for eQTLs.
a. The mean causal variant enrichment for eQTL within SCENT peaks with excluding all promoters (teal) or cis-regulatory ATAC-seq peaks with excluding all promoters (yellow) in each dataset. b. The mean causal variant enrichment for eQTL within SCENT peaks (teal) or non-SCENT peaks with matching distance to TSS (pink). c. Comparison of the mean causal variant enrichment for eQTL (y-axis) among SCENT (teal), ArchR (pink), and Signac (purple) as a function of the number of significant peak-gene pairs at each threshold of significance by FDR in SCENT and correlation r in ArchR and Signac. d. Comparison of the mean causal variant enrichment for eQTL among SCENT, ArchR, and Signac as a function of the number of significant peak-gene pairs at each threshold of FDR in SCENT, ArchR and Signac. The ArchR results with > 180,000 peak-gene linkages are omitted. e. Comparison of the mean causal variant enrichment for eQTL among SCENT, ArchR, and ArchR filtered on RNA expression as a function of the number of significant peak-gene pairs. f. Comparison of the mean causal variant enrichment for eQTL among SCENT, Signac, and Signac filtered on RNA expression as a function of the number of significant peak-gene pairs. g. Comparison of the mean causal variant enrichment for eQTL among SCENT, the default Pearson’s correlation version of Signac, and the optional Spearman’s correlation version of Signac as a function of the number of significant peak-gene pairs. h. Comparison of the mean causal variant enrichment for eQTL among original SCENT (Poisson regression + non-parametric bootstrapping), Poisson-only strategy without bootstrapping, and Cicero (correlation method using sc-ATAC-seq alone) as a function of the number of significant peak-gene pairs up to 100,000 peak-gene linkages. i. Comparison of the mean causal variant enrichment for eQTL between SCENT and Cicero peaks with adding all accessible promoter regions (1 kb regions from TSS) to account for potential promoter bias. j. Tissue-specific causal variant enrichment within SCENT peaks. The dots and lines are colored by the eQTL source tissue in GTEx that we assessed. In all panels, the bars indicate 95% confidence intervals by bootstrapping genes (nbootstrap=1000).
Extended Data Fig. 7 Causal variant enrichment for GWAS.
a and b. The mean causal variant enrichment for GWAS within cell-type-specific and aggregated SCENT enhancers (teal), ENCODE cCREs (pink), group-specific and aggregated EpiMap enhancers (red) and sample-specific and aggregated ABC enhancers (blue). GWAS results were based on FinnGen (a) and UK Biobank (b). The bars indicate 95% confidence intervals by bootstrapping traits (nbootstrap=1000). c. The mean causal variant enrichment for FinnGen GWAS (see Methods) within SCENT peaks with excluding all promoters (teal) or cis-regulatory ATAC-seq peaks with excluding all promoters (yellow) in each of the 9 single-cell datasets. The bars indicate 95% confidence intervals by bootstrapping traits (nbootstrap=1000). d. The mean causal variant enrichment for FinnGen GWAS (see Methods) within SCENT peaks (teal) or non-SCENT peaks with matching distance to TSS (pink) in each of the 9 single-cell datasets. The bars indicate 95% confidence intervals by bootstrapping traits (nbootstrap=1000). e. The fraction of known genes from Mendelian autoimmune diseases among all the genes identified by SCENT, EpiMap, and ABC model. The color of the bars indicates the cell types in each linking method.
Extended Data Fig. 8 Causal variant enrichment for GWAS and comparison with published bulk methods and single-cell methods.
a. Comparison of the mean causal variant enrichment for FinnGen GWAS (y-axis) among SCENT (teal), EpiMap (red), and ABC model (blue) as a function of the number of significant peak-gene pairs (x-axis) at each threshold of significance. The bars indicate 95% confidence intervals by bootstrapping traits (nbootstrap=1000). b. We calculated the causal variant enrichment for FinnGen GWAS among SCENT (teal), EpiMap (reds), and ABC model (blues) by changing the PIP thresholds in defining putative causal variants from fine-mapping. The bars indicate 95% confidence intervals by bootstrapping traits (nbootstrap=1000). c and d. The mean causal variant enrichment for GWAS within SCENT enhancers (teal), ArchR (pink) and Signac enhancers (purple). GWAS results were based on FinnGen (c) and UK Biobank (d) using the FDR < 10% threshold in each software and eight benchmarking datasets (see Methods). The bars indicate 95% confidence intervals by bootstrapping traits (nbootstrap=1000).
Extended Data Fig. 9 SMAD3 locus in asthma GWAS.
Rs17293632 in asthma GWAS (a) was prioritized and connected to SMAD3 gene by SCENT in myeloid cells (b). The panel a is a GWAS regional plot, with x-axis representing the position of each genetic variant and y-axis representing -log10(P) from GWAS (a two-sided P value). The rs17293632 has a significant caQTL effect, as shown in c and d. In panel c, the read coverage from single-cell ATAC-seq in each of donors with heterozygous genotype at this accessible region is presented, and at rs17293632, we observed allele-specific increased accessibility with C allele when compared T allele across donors. In panel d, normalized chromatin accessibility based on the read coverage for an individual after regressing out covariates is presented by the genotype of rs17293632 (CC, CT and TT). The horizontal bars within boxes indicate the median, and the lower and upper hinges represent 25% and 75% quantile. The upper whisker extends from the hinge to the largest value no further than 1.5 * inter-quartile range (IQR) from the hinge. The lower whisker extends from the hinge to the smallest value at most 1.5 * IQR of the hinge. All individual points are plotted as dots.
Extended Data Fig. 10 Cells to be included in the regression framework.
a. An example situation of correlated gene expression without biological regulatory function. b. Benchmarking models for statistical power to define biologically plausible peak-gene linkage over false-associations due to correlated genes. c. Benchmarking results regarding cells and covariates included in the SCENT regression model. The x-axis represents the number of statistically significant peak-gene linkages among 5,000 randomly selected peak-gene linkages in cis, and the y-axis represents the number of statistically significant peak-gene linkages in cis divided by the number of statistically significant peak-gene linkages in trans among 5,000 randomly selected peak-gene linkages on different chromosomes, as a proxy metric for capability of identifying regulatory elements over ‘correlated’ elements. Red dots indicate the analyses conducted in all cells including different cell types (n = 8,881), whereas blue dots indicate the analyses conducted in only T cells (n = 8,881). d and e. False positive rate and precision for peak-gene linkages from analyses conducted in all cells (teal) or in only T cells (orange) by using experimentally validated enhancer-gene linkages (that is, CRISPR-Flow FISH data in d and H3K27ac data in e). False negative rate and precision were defined as follows: \(false\,negative\,rate=\#\,false\,negative/(\#\,true\,positive+\#\,false\,negative)=1-recall\).
Supplementary information
Supplementary Information
Supplementary Notes 1–3 and Figs. 1–8.
Supplementary Table 1
Supplementary Tables 1–9.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sakaue, S., Weinand, K., Isaac, S. et al. Tissue-specific enhancer–gene maps from multimodal single-cell data identify causal disease alleles. Nat Genet 56, 615–626 (2024). https://doi.org/10.1038/s41588-024-01682-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-024-01682-1