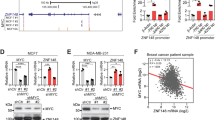
Abstract
The transcription factor MYC is overexpressed in most cancers, where it drives multiple hallmarks of cancer progression. MYC is known to promote oncogenic transcription by binding to active promoters. In addition, MYC has also been shown to invade distal enhancers when expressed at oncogenic levels, but this enhancer binding has been proposed to have low gene-regulatory potential. Here, we demonstrate that MYC directly regulates enhancer activity to promote cancer type-specific gene programs predictive of poor patient prognosis. MYC induces transcription of enhancer RNA through recruitment of RNA polymerase II (RNAPII), rather than regulating RNAPII pause-release, as is the case at promoters. This process is mediated by MYC-induced H3K9 demethylation and acetylation by GCN5, leading to enhancer-specific BRD4 recruitment through its bromodomains, which facilitates RNAPII recruitment. We propose that MYC drives prognostic cancer type-specific gene programs through induction of an enhancer-specific epigenetic switch, which can be targeted by BET and GCN5 inhibitors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD039586. Raw and processed next-generation sequencing data have been deposited to the GEO database with the dataset identifier GSE224613. Preproccesed data to re-create the figures are available on Zenodo (https://zenodo.org/records/8323614). In addition to the data generated for this study, publicly available raw sequencing data were downloaded from GEO using the following accession codes: GSE86504, GSE86511, GSE78064, GSE44672, GSE36354, GSE23893, GSE15220, GSE96867, GSE44672, GSE65201, GSE167154, GSE197062 and GSE32222. Preproccessed RNA-seq data from cell lines were downloaded from DepMap (https://depmap.org/portal/). Clinical and expression data from TCGA and METABRIC were downloaded from cBioPortal (https://www.cbioportal.org/) and Rueda et al.71. DNA binding motifs were downloaded from the JASPAR database (https://jaspar2020.genereg.net/). Source data are provided with this paper.
Code availability
The authors confirm that data processing and analysis used existing code/software packages whose details are provided in Methods. The code used for figure generation is available on GitHub (https://github.com/SimonToftholm/Jakobsen_et_al_2024).
References
Meškytė, E. M., Keskas, S. & Ciribilli, Y. MYC as a multifaceted regulator of tumor microenvironment leading to metastasis. Int. J. Mol. Sci. 21, 7710 (2020).
Rahl, P. B. et al. c-Myc regulates transcriptional pause release. Cell 141, 432–445 (2010).
Ji, H. et al. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS One 6, e26057 (2011).
Qiu, X. et al. MYC drives aggressive prostate cancer by disrupting transcriptional pause release at androgen receptor targets. Nat. Commun. 13, 2559 (2022).
Zeid, R. et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat. Genet. 50, 515–523 (2018).
Lin, C. Y. et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67 (2012).
Walz, S. et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511, 483–487 (2014).
See, Y. X. et al. MYC overexpression leads to increased chromatin interactions at superenhancers and MYC binding sites. Genome Res. 32, 629–642 (2022).
Sabò, A. et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511, 488–492 (2014).
Moore, J. E. et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020).
Bywater, M. J. et al. Reactivation of Myc transcription in the mouse heart unlocks its proliferative capacity. Nat. Commun. 11, 1827 (2020).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Corces, M. R. et al. The chromatin accessibility landscape of primary human cancers. Science 362, eaav1898 (2018).
Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Ross-Innes, C. S. et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393 (2012).
Fulco, C. P. et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669 (2019).
Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Hart, J. R. et al. Inhibitor of MYC identified in a Kröhnke pyridine library. Proc. Natl Acad. Sci. USA 111, 12556–12561 (2014).
Nie, Z. et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 (2012).
Layer, R. M. et al. GIGGLE: a search engine for large-scale integrated genome analysis. Nat. Methods 15, 123–126 (2018).
Bentsen, M. et al. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat. Commun. 11, 4267 (2020).
Conway, M. E. et al. STAT3 and GR cooperate to drive gene expression and growth of basal-like triple-negative breast cancer. Cancer Res. 80, 4355–4370 (2020).
Pan, H. et al. Forkhead box C1 boosts triple-negative breast cancer metastasis through activating the transcription of chemokine receptor-4. Cancer Sci. 109, 3794–3804 (2018).
Shlyueva, D., Stampfel, G. & Stark, A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15, 272–286 (2014).
McDowell, I. C. et al. Glucocorticoid receptor recruits to enhancers and drives activation by motif-directed binding. Genome Res. 28, 1272–1284 (2018).
West, D. C. et al. Discovery of a glucocorticoid receptor (GR) activity signature using selective GR antagonism in ER-negative breast cancer. Clin. Cancer Res. 24, 3433–3446 (2018).
Obradović, M. M. S. et al. Glucocorticoids promote breast cancer metastasis. Nature 567, 540–544 (2019).
Henriques, T. et al. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 32, 26–41 (2018).
Castell, A. et al. A selective high affinity MYC-binding compound inhibits MYC:MAX interaction and MYC-dependent tumor cell proliferation. Sci. Rep. 8, 10064 (2018).
Han, H. et al. Small-Molecule MYC Inhibitors Suppress Tumor Growth and Enhance Immunotherapy. Cancer Cell 36, 483–497.e15 (2019).
Kim, T. K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010).
Shiraki, T. et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl Acad. Sci. USA 100, 15776–15781 (2003).
Mousavi, K. et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell 51, 606–617 (2013).
Bhagwat, A. S. et al. BET bromodomain inhibition releases the mediator complex from select cis-regulatory elements. Cell Rep. 15, 519–530 (2016).
Bressin, A. et al. High-sensitive nascent transcript sequencing reveals BRD4-specific control of widespread enhancer and target gene transcription. Nat. Commun. 14, 4971 (2023).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Zhang, N. et al. MYC interacts with the human STAGA coactivator complex via multivalent contacts with the GCN5 and TRRAP subunits. Biochim. Biophys. Acta 1839, 395–405 (2014).
Jin, Q. et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 (2011).
Zippo, A. et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138, 1122–1136 (2009).
Kalkat, M. et al. MYC Protein Interactome Profiling Reveals Functionally Distinct Regions that Cooperate to Drive Tumorigenesis. Mol. Cell 72, 836–848.e7 (2018).
Majaz, S. et al. Histone acetyl transferase GCN5 promotes human hepatocellular carcinoma progression by enhancing AIB1 expression. Cell Biosci. 6, 47 (2016).
Lorenzin, F. et al. Different promoter affinities account for specificity in MYC-dependent gene regulation. eLife 5, e15161 (2016).
Andersson, R. & Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 21, 71–87 (2020).
Lourenco, C. et al. MYC protein interactors in gene transcription and cancer. Nat. Rev. Cancer 21, 579–591 (2021).
Doroshow, D. B., Eder, J. P. & LoRusso, P. M. BET inhibitors: a novel epigenetic approach. Ann. Oncol. 28, 1776–1787 (2017).
Haque, M. E. et al. The GCN5: its biological functions and therapeutic potentials. Clin. Sci. 135, 231–257 (2021).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Wu, G. et al. Menin enhances c-Myc-mediated transcription to promote cancer progression. Nat. Commun. 8, 15278 (2017).
Galardi, S. et al. Resetting cancer stem cell regulatory nodes upon MYC inhibition. EMBO Rep. 17, 1872–1889 (2016).
Xiong, X. et al. A scalable epitope tagging approach for high-throughput ChIP-seq analysis. ACS Synth. Biol. 6, 1034–1042 (2017).
Joseph, R. et al. Integrative model of genomic factors for determining binding site selection by estrogen receptor-α. Mol. Syst. Biol. 6, 456 (2010).
Wang, Y. et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 163, 174–186 (2015).
Kim, T. et al. Comparative characterization of 3D chromatin organization in triple-negative breast cancers. Exp. Mol. Med. 54, 585–600 (2022).
Castell, A. et al. MYCMI-7: a small MYC-binding compound that inhibits MYC: MAX interaction and tumor growth in a MYC-dependent manner. Cancer Res. Commun. 2, 182–201 (2022).
Franco, H. L. et al. Enhancer transcription reveals subtype-specific gene expression programs controlling breast cancer pathogenesis. Genome Res. 28, 159–170 (2018).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Papachristou, E. K. et al. A quantitative mass spectrometry-based approach to monitor the dynamics of endogenous chromatin-associated protein complexes. Nat. Commun. 9, 2311 (2018).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2016).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Fornes, O. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020).
Cvetesic, N. et al. SLIC-CAGE: high-resolution transcription start site mapping using nanogram-levels of total RNA. Genome Res. 28, 1943–1956 (2018).
Thodberg, M., Thieffry, A., Vitting-Seerup, K., Andersson, R. & Sandelin, A. CAGEfightR: analysis of 5′-end data using R/Bioconductor. BMC Bioinf. 20, 487 (2019).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Rueda, O. M. et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 567, 399–404 (2019).
Acknowledgements
Sequencing was carried out at the Center for Functional Genomics and Tissue Plasticity, in the Functional Genomics and Metabolism research unit at SDU. The authors thank T. P. Mortensen, M. Wishoff, G. Jørgensen, and R. Nielsen for sequencing assistance. Work in the R.S. laboratory was supported by the Novo Nordisk Foundation (NNF18OC0053276 and NNF21OC0071373 to R.S.), the Danish Cancer Society (R231-A13786 to R.S.), and the VILLUM Foundation (project no. 42066 to R.S.). The mass spectrometry and proteomics platform at SDU, used to perform qPLEX-RIME, was financed by grants from the VILLUM Foundation (VILLUM Center for Bioanalytical Sciences, grant no. 7292 to O.N.J.), the Novo Nordisk Foundation (INTEGRA grant no. NNF20OC0061575 to O.N.J.), and by PRO-MS: Danish National Mass Spectrometry Platform for Functional Proteomics (grant no. 5072-00007B to O.N.J.). The work in the R.A. laboratory was supported by the Novo Nordisk Foundation (NNF20OC0059796 and NNF21SA0072102 to R.A.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
S.T.J. and R.S. conceived and designed the study. S.T.J., R.A.M.J., M.S.M., T.R., C.S.V. and M.S.S. performed the experiments. S.T.J., T.R. and H.E. performed the computational analysis. R.S., O.N.J. and R.A. supervised the study. S.T.J. and R.S. wrote the manuscript with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Catherine Wilson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 MYC expression and chromatin binding.
a, MYC expression across cancer cell lines summarized for tumor types from depmap.org. Boxplot: Center line, median; box limits; upper and lower quartiles; whiskers, 1,5x interquartile range; points, outliers. b, MYC expression in four breast cancer cell lines from depmap.org. c, Occurrence of strong/weak E-box motifs within MYC-bound promoters (−1 kb to +100 bp around the TSS) and enhancers (intronic and intergenic regions) from BT549, MDA-MB-231, MCF7, and T47D cell lines. d, Average MYC ChIP-seq signal at MYC-bound promoters and enhancers across four breast cancer cell lines. e, Spearman correlation of MYC binding across four breast cancer cell lines in MYC-bound enhancers (n = 89,631) and promoters (n = 10,049) called in at least one of the cell lines. Circle size and color indicate the correlation values (range: 0 to 1). f, Genome browser screenshot of MYC breast cancer subtype-specific enhancers defined in Fig. 1d. g, Accessibility of MYC breast cancer subtype-specific enhancers from Fig. 1d is shown for TNBC (n = 28) and ER+ (n = 109) breast cancer patients based on the TCGA ATAC-seq data13. Boxplots show the average accessibility of the given regions for patients in each group. P values were calculated using Welch’s two-sided t-test. Boxplot: Center line, median; box limits; upper and lower quartiles; whiskers, 1,5x interquartile range; outliers are not shown. h, Expression of genes located within 50 kb from any of the breast cancer subtype-specific enhancers defined in Fig. 1d in TNBC and ER+ breast cancers from TCGA14. Average expression of the gene groups for each patient (TNBC n = 292, ER+ n = 867) is summarized. P values were calculated using Welch’s two-sided t-test. Boxplot: Center line, median; box limits; upper and lower quartiles; whiskers, 1,5x interquartile range; outliers are not shown. i, Differential binding of MYC in response to 1 h fulvestrant treatment (100 nM) in MCF7 cells as determined by ChIP-seq. Red dots indicate significantly regulated binding sites (Padj < 0.05, two sided, FDR corrected using the Benjamini–Hochberg method in DiffBind). j, Protein expression of ER and MYC during 1 h fulvestrant treatment (100 nM) in MCF7 cells. β-tubulin is included as a control. The blot is representative of three independent biological replicates.
Extended Data Fig. 2 MYC enhancer and promoter binding target different gene programs.
a, Same as in Fig. 2a for MCF7 cells considering all genes within 50 kb of MYC binding sites. Lines represent the means and gray ranges represent 95% CI. b, Density of enhancer–promoter pairs involving MYC-bound enhancers over the promoter/enhancer ranking of putative MYC target genes in BT549 cells from Fig. 2a. Enhancer–promoter pairs were extracted from17. Line represents the mean and gray range represents 95% CI. Statistical testing was performed using one-way analysis of variance (ANOVA). c, Western blot validation of 72 h siRNA-mediated MYC knockdown and/or 2 h Dexamethasone (Dex) treatment in BT549 cells. β-tubulin is included as a control. Results from three independent biological replicates (Rep) are shown.
Extended Data Fig. 3 Validation of KJ-PYR-9.
a, Genome browser screenshot showing MYC ChIP-seq in response to KJ-PYR-9 treatment (3 h) for part of chromosome 8. The y-axis shows normalized FPKM counts. b, Differentially expressed genes (Padj < 0.05, two-sided, FDR corrected using the Benjamini–Hochberg method in DEseq2) upon KJ-PYR-9 treatment (3 h) in BT549 cells were identified and their response to KJ-PYR-9 was compared with the response to siRNA-mediated knockdown of MYC in a scatterplot. The color intensity indicates the density of genes in the scatterplot. c, Enrichment of three MYC target gene sets over genes ranked by Log2 fold change (Log2FC) in response to KJ-PYR-9 treatment (3 h) (part of the data from Fig. 4f) or siRNA-mediated MYC knockdown in BT549 cells. d, Percentage of ERCC spike-ins in RNA-seq libraries from BT549 cells upon siRNA-mediated knockdown of MYC or KJ-PYR-9 treatment (3 h) (part of the data from Fig. 4f) in three independent biological replicates. Bars represent mean and error bars represent s.e.m. P values were determined by paired Student’s t-test (two-sided).
Extended Data Fig. 4 Crosstalk between MYC and cancer type-specific transcription factors at enhancers.
a, Enrichment of a weak E-box motif (Extended Data Fig. 1c) in MYC-bound enhancers significantly activated or constitutively active as determined by MED1 ChIP-seq in response to KJ-PYR-9 treatment (3 h) in BT549 cells (see Fig. 3c). b, Average MYC ChIP-seq signal in BT549 cells at MYC-activated, MYC-repressed, and constitutively active enhancers as determined by MED1 ChIP-seq (see Fig. 3c). c, Percentage overlap of MYC enhancers and promoters with differential MED1 binding upon KJ-PYR-9 treatment (3 h) defined in Fig. 3b,c between BT549 and MCF7 cells. d, Giggle analysis22 on MYC-activated enhancers from MCF7 and BT549 cells (Fig. 3c). Bars indicate the mean of the observed Giggle scores and dots represent individual datasets. A high Giggle score indicates a high overlap with chromatin binding of a given transcription factor as determined by ChIP-seq. Only ChIP-seq experiments performed in TNBC or ER+ breast cancer cell lines were used for this analysis. e, Change in STAT3 footprint scores calculated from corrected cut counts within MYC-bound promoters (n = 3811) and enhancers (n = 6723) upon KJ-PYR-9 treatment (3 h) in BT549 cells. Boxplot: Center line, median; box limits; upper and lower quartiles; whiskers, 1,5x interquartile range; outliers are not shown. P value was calculated with Welch’s t-test, two-sided. f, MED1 binding in MCF7 cells upon KJ-PYR-9 treatment (3 h) at ER-bound MYC enhancers (see Fig. 1f).
Extended Data Fig. 5 Co-binding of MYC and GR on chromatin.
ChIP-seq heatmaps (left) and average line plots (right) for all common GR and MYC enhancer binding sites upon Dex treatment (2 h) in BT549 cells. GR Tag counts were normalized to a spike-in control.
Extended Data Fig. 6 MYC-mediated regulation of enhancer features.
a, Same as Fig. 5a for genes that are not regulated upon 3 h KJ-PYR-9 treatment (Padj > 0.2, two-sided, FDR corrected using the Benjamini–Hochberg method in DEseq2) without strong MYC binding at the promoter (Tag counts < 25) in BT549 cells. P value was calculated using Welch’s two-sided t-test. b, Average spike-in normalized MYC ChIP-seq signal in response to MYCMI-6 and MYCi975 treatment for 3 h in BT549 cells. The average signal for all MYC-bound enhancers and promoters is shown. c, Average ChIP-seq signal for total RNAPII at MYC-activated enhancers located within introns or intergenic regions in response to KJ-PYR-9 treatment (3 h) of BT549 cells. d, Changes in CAGE-seq signal in response to 3 h KJ-PYR-9 treatment in BT549 cells are plotted for MYC-activated, constitutive, and MYC-repressed enhancers as determined by changes in MED1 binding during KJ-PYR-9 treatment (see Fig. 3c). * P < 0.05 calculated using Welch’s two-sided t-test. e, eRNA within MYC-activated enhancers not located within genes (n = 1899) measured using rRNA-depleted (Ribo-Zero) RNA-seq in BT549 cells (part of the data from Fig. 4f). Boxplot: Center line, median; box limits; upper and lower quartiles; whiskers, 1,5x interquartile range; outliers are not shown. P value was calculated using Welch’s two-sided t-test. f, Heatmaps showing ATAC-seq and MED1 ChIP-seq signal at MYC-activated enhancers (see Fig. 3c) in response to KJ-PYR-9 treatment (3 h) in BT549 cells.
Extended Data Fig. 7 Loss of BRD4 binding at enhancers upon MYC inhibition.
a, Average BRD4 ChIP-seq signal at MYC-activated enhancers and gene promoters (see Figs. 3c and 5a) in response to MYCMI-6 and MYCi975 treatment (3 h) in BT549 cells. b, Western blot for BRD4 in response to KJ-PYR-9 treatment (3 h) in BT549 cells. β-tubulin is included as a control. Representative of two independent biological replicates. c, Western blot of BRD4 in response to MYCMI-6 and MYCi975 treatment (3 h) in BT549 cells. β-tubulin is included as a control. Representative of three independent biological replicates. d, Western blot showing BRD4, GCN5, MYC, and H3K9ac levels upon JQ1 treatment (1 h) in BT549 cells. β-tubulin is included as a control. Representative of two independent biological replicates.
Extended Data Fig. 8 Loss of GCN5, H3K9ac, and KDM3A at enhancers upon MYC inhibition.
a, Venn diagram showing overlap between GCN5 (veh) and MYC (from Fig. 1a) binding as determined by ChIP-seq in BT549 cells (top). Genomic annotation of shared GCN5-MYC binding sites (bottom). b, Average GCN5 ChIP-seq signal at MYC-bound promoters and enhancers in BT549 cells. c, Western blot showing KDM3A, GCN5, MYC, H3K9me1/2, and H3K9ac protein levels upon KJ-PYR-9 treatment (3 h) in BT549 cells. β-Tubulin and H3 are included as controls. Representative of three independent biological replicates. d, Western blot showing GCN5, MYC, and H3K9ac upon MYCi975 treatment (3 h) in BT549 cells. β-Tubulin and H3 are included as controls. Representative of three independent biological replicates. Vertical black line indicates that lanes between the two conditions were cropped. e and f, Change in GCN5 binding at all MYC-bound promoters and enhancers in response to 3 h KJ-PYR-9 (e) or MYCi975 (f) treatment of BT549 cells. Black lines in the horizontal violin plots represent the median. P values were calculated using Welch’s two-sided t-test. g, Average ChIP-seq signal for GCN5 and H3K9ac at MYC-activated enhancers and gene promoters (defined in Figs. 3c and 5a) in response to MYCi975 treatment (3 h) of BT549 cells. h, Boxplot showing the change in KDM3A binding with KJ-PYR-9 treatment (3 h) as determined by ChIP-seq in BT549 cells at shared (KDM3A and MYC) (enhancers n = 10327 and promoters n = 7094) or KDM3A-only (enhancers n = 6320 and promoters n = 2138) enhancers and promoters. Boxplot: Center line, median; box limits; upper and lower quartiles; whiskers, 1,5x interquartile range; outliers are not shown. P values were calculated using Welch’s t-test, two-sided. i, Venn diagram showing overlap between KDM3A and MYC binding sites as determined by ChIP-seq in BT549 cells. The percentages of shared MYC-KDM3A binding sites in relevant sub-groups of the Venn are indicated. j, Western blot showing BRD4, GCN5, MYC, and H3K9ac levels in response to siRNA-mediated GCN5 knockdown in BT549 cells. β-tubulin is included as a control. Representative of two independent biological replicates. k, BT549 cell growth upon siRNA-mediated knockdown of GCN5 as determined by Incucyte live-cell imaging. The lines show the average confluency of 15 technical replicates and the error bars represent s.e.m. The data shown is representative of two independent biological replicates.
Supplementary information
Source data
Source Data Extended Data Figs. 1, 2, 7 and 8
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jakobsen, S.T., Jensen, R.A.M., Madsen, M.S. et al. MYC activity at enhancers drives prognostic transcriptional programs through an epigenetic switch. Nat Genet 56, 663–674 (2024). https://doi.org/10.1038/s41588-024-01676-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-024-01676-z