Abstract
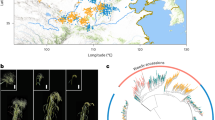
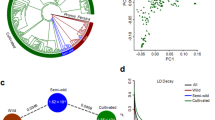
Peanut (Arachis hypogaea L.) is an important allotetraploid oil and food legume crop. China is one of the world’s largest peanut producers and consumers. However, genomic variations underlying the migration and divergence of peanuts in China remain unclear. Here we reported a genome-wide variation map based on the resequencing of 390 peanut accessions, suggesting that peanuts might have been introduced into southern and northern China separately, forming two cultivation centers. Selective sweep analysis highlights asymmetric selection between the two subgenomes during peanut improvement. A classical pedigree from South China offers a context for the examination of the impact of artificial selection on peanut genome. Genome-wide association studies identified 22,309 significant associations with 28 agronomic traits, including candidate genes for plant architecture and oil biosynthesis. Our findings shed light on peanut migration and diversity in China and provide valuable genomic resources for peanut improvement.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All the 390 genomic sequence data for GWAS analysis have been deposited in the National Center for Biotechnology Information (NCBI) database under BioProject number PRJNA776707. All the 11 varieties of genomic sequence data for IBD analysis have been deposited in the NCBI database under BioProject number PRJNA1031811. The SNP and InDel genotypes have been deposited in Zenodo81 (https://doi.org/10.5281/zenodo.10054109). The published transcriptomic datasets for candidate gene expression analysis can be downloaded from the NCBI Sequence Read Archive under accession numbers SRP167797 and SRP033292 mentioned in the corresponding original literature. Source data are provided with this paper.
Code availability
Custom scripts for calculating the coverage of the aligned sequence are available at Zenodo82 (https://doi.org/10.5281/zenodo.10023694).
References
Akram, N. A., Shafiq, F. & Ashraf, M. Peanut (Arachis hypogaea L.): a prospective legume crop to offer multiple health benefits under changing climate. Compr. Rev. Food Sci. Food Saf. 17, 1325–1338 (2018).
Fávero, A. P., Simpson, C. E., Valls, J. M. & Velo, N. A. Study of evolution of cultivated peanut through crossability studies among Arachis ipaënsis, A. duranensis, and A. hypogaea. Crop Sci. 46, 1546–1552 (2006).
Seijo, G. et al. Genomic relationships between the cultivated peanut (Arachis hypogaea, Leguminosae) and its close relatives revealed by double GISH. Am. J. Bot. 94, 1963–1971 (2007).
Samoluk, S. S. et al. First insight into divergence, representation and chromosome distribution of reverse transcriptase fragments from L1 retrotransposons in peanut and wild relative species. Genetica 143, 113–125 (2015).
Bertioli, D. J. et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446 (2016).
Bertioli, D. J. et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51, 877–884 (2019).
Zhuang, W. et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 51, 865–876 (2019).
Yin, D. et al. Genome of an allotetraploid wild peanut Arachis monticola: a de novo assembly. GigaScience. 7, giy066 (2018).
Pandey, M. K. et al. Advances in Arachis genomics for peanut improvement. Biotechnol. Adv. 30, 639–651 (2012).
Li, L. et al. GWAS and bulked segregant analysis reveal the loci controlling growth habit‑related traits in cultivated peanut (Arachis hypogaea L.). BMC Genomics 23, 403 (2022).
Li, L. et al. Construction of high-density genetic map and mapping quantitative trait loci for growth habit-related traits of peanut (Arachis hypogaea L.). Front. Plant Sci. 10, 745 (2019).
Luo, H. et al. Next-generation sequencing identified genomic region and diagnostic markers for resistance to bacterial wilt on chromosome B02 in peanut (Arachis hypogaea L.). Plant Biotechnol. J. 17, 2356–2369 (2019).
Zhao, K. et al. PSW1, an LRR receptor kinase, regulates pod size in peanut. Plant Biotechnol. J. 21, 2113–2124 (2023).
Han, S. et al. AhNPR3 regulates the expression of WRKY and PR genes, and mediates the immune response of the peanut (Arachis hypogaea L.). Plant J. 110, 735–747 (2022).
Lu, Q. et al. Consensus map integration and QTL meta-analysis narrowed a locus for yield traits to 0.7 cM and refined a region for late leaf spot resistance traits to 0.38 cM on linkage group A05 in peanut (Arachis hypogaea L.). BMC Genomics 19, 887 (2018).
Luo, H. et al. Discovery of genomic regions and candidate genes controlling shelling percentage using QTL-seq approach in cultivated peanut (Arachis hypogaea L.). Plant Biotechnol. J. 17, 1248–1260 (2019).
Yang, Y. et al. Genetic analysis and exploration of major effect QTLs underlying oil content in peanut. Theor. Appl. Genet. 136, 97 (2023).
Zhu, C., Gore, M., Buckler, E. S. & Yu, J. Status and prospects of association mapping in plants. Plant Genome 1, 5–20 (2008).
Huang, X. et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967 (2010).
Huang, X. et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501 (2012).
Tian, F. et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 43, 159–162 (2011).
Zhou, Z. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 33, 408–414 (2015).
Varshney, R. K. et al. Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat. Genet. 51, 857–864 (2019).
Varshney, R. K. et al. A chickpea genetic variation map based on the sequencing of 3,366 genomes. Nature 599, 622–627 (2021).
Fan, W. et al. Sequencing of Chinese castor lines reveals genetic signatures of selection and yield-associated loci. Nat. Commun. 10, 3418 (2019).
Jia, G. et al. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 45, 957–961 (2013).
Fang, L. et al. Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat. Genet. 49, 1089–1098 (2017).
Ma, Z. et al. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 50, 803–813 (2018).
Kang, L. et al. Genomic insights into the origin, domestication and diversification of Brassica juncea. Nat. Genet. 53, 1392–1402 (2021).
Lu, K. et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 10, 1154 (2019).
Guo, J. et al. Association of yield-related traits in founder genotypes and derivatives of common wheat (Triticum aestivum L.). BMC Plant Biol. 18, 38 (2018).
Zhang, X. et al. Genome-wide association study of major agronomic traits related to domestication in peanut. Front. Plant Sci. 8, 1611 (2017).
Liu, Y. et al. Genomic insights into the genetic signatures of selection and seed trait loci in cultivated peanut. J. Adv. Res. 42, 237–248 (2022).
Chen, X. et al. Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant 12, 920–934 (2019).
Chen, X. et al. Draft genome of the peanut A-genome progenitor (Arachis duranensis) provides insights into geocarpy, oil biosynthesis, and allergens. Proc. Natl Acad. Sci. USA 113, 6785–6790 (2016).
Pickrell, J. K. & Pritchard, J. K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8, e1002967 (2012).
Collin, F. D. et al. Extending approximate Bayesian computation with supervised machine learning to infer demographic history from genetic polymorphisms using DIYABC Random Forest. Mol. Ecol. Resour. 21, 2598–2613 (2021).
Pandey, M. K. et al. Identification of QTLs associated with oil content and mapping FAD2 genes and their relative contribution to oil quality in peanut (Arachis hypogaea L.). BMC Genet. 15, 133 (2014).
Zhao, Y. et al. Whole-genome resequencing-based QTL-seq identified AhTc1 gene encoding a R2R3-MYB transcription factor controlling peanut purple testa colour. Plant Biotechnol. J. 18, 96–105 (2020).
Chen, H., Patterson, N. & Reich, D. Population differentiation as a test for selective sweeps. Genome Res. 20, 393–402 (2010).
Gangurde, S. S. et al. Nested-association mapping (NAM)-based genetic dissection uncovers candidate genes for seed and pod weights in peanut (Arachis hypogaea). Plant Biotechnol. J. 18, 1457–1471 (2020).
Meng, L. S., Wang, Z. B., Yao, S. Q. & Liu, A. The ARF2-ANT-COR15A gene cascade regulates ABA-signaling-mediated resistance of large seeds to drought in Arabidopsis. J. Cell Sci. 128, 3922–3932 (2015).
Schruff, M. C. et al. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133, 251–261 (2006).
Okamuro, J. K., Caster, B., Villarroel, R., Van Montagu, M. & Jofuku, K. D. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl Acad. Sci. USA 94, 7076–7081 (1997).
Zhao, M. et al. DROOPY LEAF1 controls leaf architecture by orchestrating early brassinosteroid signaling. Proc. Natl Acad. Sci. USA 117, 21766–21774 (2020).
Sreeramulu, S. et al. BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 74, 905–919 (2013).
Kong, Q., Yuan, L. & Ma, W. WRINKLED1, a ‘Master Regulator’ in transcriptional control of plant oil biosynthesis. Plants (Basel) 8, 238 (2019).
Li, Q. et al. Wrinkled1 accelerates flowering and regulates lipid homeostasis between oil accumulation and membrane lipid anabolism in Brassica napus. Front. Plant Sci. 6, 1015 (2015).
Liu, J. et al. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 48, 9–15 (2010).
Chen, B. et al. Multiple GmWRI1s are redundantly involved in seed filling and nodulation by regulating plastidic glycolysis, lipid biosynthesis and hormone signalling in soybean (Glycine max). Plant Biotechnol. J. 18, 155–171 (2020).
Pouvreau, B. et al. Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 156, 674–686 (2011).
Tyczewska, A., Woźniak, E., Gracz, J., Kuczyński, J. & Twardowski, T. Towards food security: current state and future prospects of agrobiotechnology. Trends Biotechnol. 36, 1219–1229 (2018).
Moretzsohn, M. et al. Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome. BMC Plant Biol. 4, 11 (2004).
Ferguson, M. E., Bramel, P. J. & Chandra, S. Gene diversity among botanical varieties in peanut (Arachis hypogaea L.). Crop Sci. 44, 1847–1854 (2004).
Khera, P. et al. Single nucleotide polymorphism-based genetic diversity in the reference set of peanut (Arachis spp.) by developing and applying cost-effective kompetitive allele specific polymerase chain reaction genotyping assays. Plant Genome 6, (2013).
Wang, H. et al. Analysis of genetic diversity and population structure of peanut cultivars and breeding lines from China, India and the US using simple sequence repeat markers. J. Integr. Plant Biol. 58, 452–465 (2016).
Shi, H. et al. BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25, 1143–1157 (2013).
Qu, J. et al. Dissecting functions of KATANIN and WRINKLED1 in cotton fiber development by virus-induced gene silencing. Plant Physiol. 160, 738–748 (2012).
Liu, Z. J. et al. Over-expression of transcription factor GhWRI1 in upland cotton. Biol. Plant. 62, 335–342 (2018).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Vilella, A. J. et al. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19, 327–335 (2009).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Zhang, C., Dong, S. S., Xu, J. Y., He, W. M. & Yang, T. L. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 35, 1786–1788 (2019).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Kang, H. M. et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354 (2010).
Moran, M. D. Arguments for rejecting the sequential Bonferroni in ecological studies. Oikos 100, 403–405 (2003).
Shin, J. H., Blay, S., McNeney, B. & Graham, J. LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Softw. 16, Code Snippet 3 (2006).
Chen, C. et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202 (2020).
Chen, X. et al. Transcriptome-wide sequencing provides insights into geocarpy in peanut (Arachis hypogaea L.). Plant Biotechnol. J. 14, 1215–1224 (2016).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45 (2001).
Untergasser, A. et al. Primer3-new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Ahlmann-Eltze, C. & Patil, I. ggsignif: R package for displaying significance brackets for ‘ggplot2’. PsyArXiv https://doi.org/10.31234/osf.io/7awm6 (2021).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag: 2016).
Lu, Q. SNPs and InDels identified in 390 peanut accessions. Zenodo https://doi.org/10.5281/zenodo.10054109 (2023).
Lu, Q. An in-house Perl script used for the calculation of the coverage of aligned sequences (1.0). Zenodo https://doi.org/10.5281/zenodo.10023694 (2023).
Acknowledgements
This research was partially supported by the Open Competition Program of Top 10 Critical Priorities of Agricultural Science and Technology Innovation for the 14th Five-Year Plan in Guangdong Province (2022SDZG05 to X.C.), the National Natural Science Foundation of China (32301869 to L.H. and 32172051 to Q.L.), the China Agriculture Research System of MOF and MARA (CARS-13 to X.L.), the Guangdong Provincial Key Research and Development Program-Modern Seed Industry (2020B020219003 to X.C. and 2022B0202060004 to Y.H.), the Guangdong Basic and Applied Basic Research Foundation (2023A1515010098 and 2021A1515010811 to Q.L.), the Guangdong Provincial Department of Science and Technology Project-International Scientific and Technological Cooperation (20200503 to Y.H.), the Special Support Program of Guangdong Province (2021TX06N789 to X.C.), the Agricultural Competitive Industry Discipline Team Building Project of Guangdong Academy of Agricultural Sciences (202104TD to X.C.), the Special Fund for Scientific Innovation Strategy-Construction of High Level Academy of Agriculture Science (R2020PY-JX004 to Q.L., R2020PY-JG005 to X.C. and R2021PY-QY003 to Hao Liu), the Open Fund of Guangdong Provincial Key Laboratory of Crop Genetic Improvement (202101 to Hao Liu and 202201 to Q.L.) and Start-Up grant to R.K.V.
Author information
Authors and Affiliations
Contributions
Q.L., X.C., Y.H., X.L. and R.K.V. conceived and designed the study. Q.L., Hao Liu, H. Li, D.G., L.H. and S.L. performed data analysis. Haiyan Liu and R.W. prepared the samples. Q.D. and P.D. measured the agronomic traits. Q.L. wrote the manuscript. R.K.V., V.G., A.C., M.K.P. and S.S.G. revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Hon-Ming Lam, Eric von Wettberg, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–26.
Supplementary Tables
Supplementary Tables 1–22.
Source data
Source Data Fig. 1
Source data for Fig. 1a–e,g.
Source Data Fig. 2
Source data for Fig. 2c,d.
Source Data Fig. 3
Source data for Fig. 3i,j.
Source Data Fig. 4
Source data for Fig. 4b,h,i.
Source Data Fig. 5
Source data for Fig. 5d,g–i,l–q,s,u–w,x.
Source Data Fig. 6
Source data for Fig. 6e,h–l,n–v.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, Q., Huang, L., Liu, H. et al. A genomic variation map provides insights into peanut diversity in China and associations with 28 agronomic traits. Nat Genet 56, 530–540 (2024). https://doi.org/10.1038/s41588-024-01660-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-024-01660-7