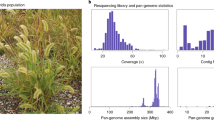
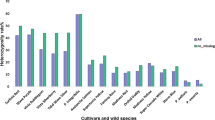
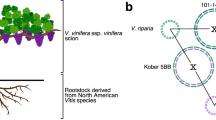
Abstract
Dwarfing rootstocks have transformed the production of cultivated apples; however, the genetic basis of rootstock-induced dwarfing remains largely unclear. We have assembled chromosome-level, near-gapless and haplotype-resolved genomes for the popular dwarfing rootstock ‘M9’, the semi-vigorous rootstock ‘MM106’ and ‘Fuji’, one of the most commonly grown apple cultivars. The apple orthologue of auxin response factor 3 (MdARF3) is in the Dw1 region of ‘M9’, the major locus for rootstock-induced dwarfing. Comparing ‘M9’ and ‘MM106’ genomes revealed a 9,723-bp allele-specific long terminal repeat retrotransposon/gypsy insertion, DwTE, located upstream of MdARF3. DwTE is cosegregated with the dwarfing trait in two segregating populations, suggesting its prospective utility in future dwarfing rootstock breeding. In addition, our pipeline discovered mobile mRNAs that may contribute to the development of dwarfed scion architecture. Our research provides valuable genomic resources and applicable methodology, which have the potential to accelerate breeding dwarfing rootstocks for apple and other perennial woody fruit trees.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All raw sequencing reads of PacBio HiFi, Hi-C, Illumina, Iso-Seq and RNA-seq have been deposited in the NCBI database with the BioProject PRJNA814760 (Supplementary Table 32). Genome assemblies have been deposited in the NCBI under accession JAXBLQ000000000 (Fuji haploid consensus genome), JAXBLN000000000 (M9 haploid consensus genome) and JAXBLK000000000 (MM106 haploid consensus genome) for the haploid consensus genome and JAXBLO000000000 (Fuji haplome A genome), JAXBLP000000000 (Fuji haplome B genome), JAXBLL000000000 (M9 haplome A genome), JAXBLM000000000 (M9 haplome B genome), JAXBLI000000000 (MM106 haplome A genome) and JAXBLJ000000000 (MM106 haplome B genome) for the haplotype-resolved genome. The assembly and annotation files have also been submitted to the National Genomics Data Center (https://ngdc.cncb.ac.cn/) with the BioProject number PRJCA010465. The genome assembly and annotation files are also available at Figshare (https://figshare.com/projects/Near-gapless_and_haplotype-resolved_apple_genomes_provide_insights_into_the_genetic_basis_of_rootstock-induced_dwarfing/190926). Source data are provided with this paper.
Code availability
Computational pipelines related to identification of mobile mRNAs can be accessed through GitHub (https://github.com/simoncchu/RNAGlass) and Zenodo (https://zenodo.org/records/10146626)98.
References
Pingali, P. L. Green revolution: impacts, limits, and the path ahead. Proc. Natl Acad. Sci. USA 109, 12302–12308 (2012).
Boss, P. K. & Thomas, M. R. Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature 416, 847–850 (2002).
McClymont, L., Goodwin, I., Whitfield, D., O’Connell, M. & Turpin, S. Effects of rootstock, tree density and training system on early growth, yield and fruit quality of blush pear. HortScience 56, 1408–1415 (2021).
Habibi, F., Liu, T., Folta, K. & Sarkhosh, A. Physiological, biochemical, and molecular aspects of grafting in fruit trees. Hortic. Res. 9, uhac032 (2022).
Ou, C. et al. A de novo genome assembly of the dwarfing pear rootstock Zhongai 1. Sci. Data 6, 281 (2019).
Prassinos, C. et al. Rootstock-induced dwarfing in cherries is caused by differential cessation of terminal meristem growth and is triggered by rootstock-specific gene regulation. Tree Physiol. 29, 927–936 (2009).
Hatton, R. G. Paradise apple stocks their fruit and blossom described. J. R. Hortic. Soc. 44, 89–94 (1919).
Foster, T. M., McAtee, P. A., Waite, C. N., Boldingh, H. L. & McGhie, T. K. Apple dwarfing rootstocks exhibit an imbalance in carbohydrate allocation and reduced cell growth and metabolism. Hortic. Res. 4, 17009 (2017).
Wang, Y. et al. Progress of apple rootstock breeding and its use. Hortic. Plant J. 5, 183–191 (2019).
MM.106. Department of Primary Industries www.dpi.nsw.gov.au/agriculture/horticulture/pomes/apples/rootstock/mm106 (2023).
Foster, T. M., Celton, J., Chagné, D., Tustin, D. S. & Gardiner, S. E. Two quantitative trait loci, Dw1 and Dw2, are primarily responsible for rootstock-induced dwarfing in apple. Hortic. Res. 2, 15001 (2015).
Fazio, G. et al. Dw2, a new dwarfing locus in apple rootstocks and its relationship to induction of early bearing in apple scions. J. Am. Soc. Hortic. 139, 87–98 (2014).
Harrison, N. et al. A new three-locus model for rootstock-induced dwarfing in apple revealed by genetic mapping of root bark percentage. J. Exp. Bot. 67, 1871–1881 (2016).
Gardner, K. M. et al. Fast and cost-effective genetic mapping in apple using next-generation sequencing. G3 (Bethesda) 4, 1681–1687 (2014).
Daccord, N. et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 49, 1099–1106 (2017).
Zhang, L. et al. A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 10, 1494 (2019).
Zhou, Q. et al. Haplotype-resolved genome analyses of a heterozygous diploid potato. Nat. Genet. 52, 1018–1023 (2020).
Sun, X. et al. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 52, 1423–1432 (2020).
Khan, A. et al. A phased, chromosome-scale genome of ‘Honeycrisp’ apple (Malus domestica). GigaByte 2022, gigabyte69 (2022).
Wang, J., Jiang, L. & Wu, R. Plant grafting: how genetic exchange promotes vascular reconnection. New Phytol. 214, 56–65 (2017).
Christoph, J. T. et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 1, 15025 (2015).
Duan, X. et al. PbWoxT1 mRNA from pear (Pyrus betulaefolia) undergoes long-distance transport assisted by a polypyrimidine tract binding protein. New Phytol. 210, 511–524 (2016).
Liu, W. et al. Identification of long-distance transmissible mRNA between scion and rootstock in cucurbit seedling heterografts. Int. J. Mol. Sci. 21, 5253 (2020).
Wang, Y. et al. A universal pipeline for mobile mRNA detection and insights into heterografting advantages under chilling stress. Hortic. Res. 7, 13 (2020).
Yang, Y. et al. Messenger RNA exchange between scions and rootstocks in grafted grapevines. BMC Plant Biol. 15, 251 (2015).
Gupta, I. et al. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat. Biotechnol. 36, 1197–1202 (2018).
Zhu, C. et al. Single-molecule, full-length transcript isoform sequencing reveals disease-associated RNA isoforms in cardiomyocytes. Nat. Commun. 12, 4203 (2021).
Xue, H. et al. Chromosome level high-density integrated genetic maps improve the Pyrus bretschneideri ‘DangshanSuli’ v1.0 genome. BMC Genomics 19, 833 (2018).
Linsmith, G. et al. Pseudo-chromosome-length genome assembly of a double haploid ‘Bartlett’ pear (Pyrus communis L.). GigaScience 8, 12 (2019).
Verde, I. et al. The Peach v2.0 release: high-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genomics 18, 225 (2017).
Jiang, F. C. et al. The apricot (Prunus armeniaca L.) genome elucidates Rosaceae evolution and β-carotenoid synthesis. Hortic. Res. 6, 128 (2019).
Li, Y. et al. Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic. Res. 6, 61 (2019).
Shao, K. J., Li, D. K. & Zhang, Z. R. A study on the breeding of apple dwarfing rootstock SH series. Acta Agric. Boreal. Sin. 3, 86–93 (1988).
Shao, K. J., Li, D. K., Zhang, Z. R. & Gao, X. M. Study on the characters and physiological trait of SH series apple draft stocks. Acta Hortic. Sin. 18, 289–295 (1991).
Cheng, H. et al. Haplotype-resolved assembly of diploid genomes without parental data. Nat. Biotechnol. 40, 1332–1335 (2022).
Butelli, E. et al. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24, 1242–1255 (2012).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Dixon, J. R. et al. Chromatin domains: the unit of chromosome organization. Mol. Cell 62, 668–680 (2016).
Melo, U. S. et al. Hi-C identifies complex genomic rearrangements and TAD-shuffling in developmental diseases. Am. J. Hum. Genet. 106, 872–884 (2020).
Long, Y. et al. Disruption of topologically associating domains by structural variations in tetraploid cottons. Genomics 113, 3405–3414 (2021).
Zhai, L. et al. Molecular and physiological characterization of the effects of auxin-enriched rootstock on grafting. Hortic. Res. 8, 74 (2021).
Fazio, G., Robinson, T. L. & Aldwinckle, H. S. The Geneva apple rootstock breeding program. Plant Breed. Rev. 39, 379–424 (2015).
Seleznyova, A. N., Tustin, D. S., White, M. D. & Costes, E. Analysis of the earliest-observed expression of dwarfing rootstock effects on young apple trees, using application of Markovian models. Acta Hortic. 732, 79–84 (2007).
Seleznyova, A., Tustin, D. S. & Thorp, T. G. Apple dwarfing rootstocks and interstocks affect the type of growth units produced during the annual growth cycle: precocious transition to flowering affects the composition and vigour of annual shoots. Ann. Bot. 101, 679–687 (2008).
Song, C. et al. Expression analysis of key auxin synthesis, transport, and metabolism genes in different young dwarfing apple trees. Acta Physiol. Plant 38, 43 (2016).
Sessions, R. A. & Zambryski, P. C. Arabidopsis gynoecium structure in the wild and in ettin mutants. Development 121, 1519–1532 (1995).
Nemhauser, J. L., Feldman, L. J. & Zambyrski, P. C. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127, 3877–3888 (2000).
Pekker, I., Alvarez, J. P. & Eshed, Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17, 2899–2910 (2005).
Izhaki, A. & Bowman, J. L. KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19, 495–508 (2007).
Ilegems, M. et al. Interplay of auxin, KANADI and class III HD-ZIP transcription factors in vascular tissue formation. Development 137, 975–984 (2010).
Kelley, D. R., Arreola, A., Gallagher, T. L. & Gasser, C. S. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139, 1105–1109 (2012).
Kuhn, A. et al. Direct ETTIN-auxin interaction controls chromatin states in gynoecium development. eLife 9, e51787 (2020).
Liu, X. et al. Auxin response factor 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. 80, 629–641 (2014).
Zhang, K. et al. Cell- and noncell-autonomous auxin response factor 3 controls meristem proliferation and phyllotactic patterns. Plant Physiol. 190, 2335–2349 (2022).
Chuong, E. B., Elde, N. C. & Feschotte, C. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18, 71–86 (2017).
Dubin, M. J., Mittelsten Scheid, O. & Becker, C. Transposons: a blessing curse. Curr. Opin. Plant Biol. 42, 23–29 (2018).
Meinke, D. W. A survey of dominant mutations in Arabidopsis thaliana. Trends Plant Sci. 18, 84–91 (2013).
Li, J., Dai, X. & Zhao, Y. A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiol. 140, 899–908 (2006).
Tworkoski, T. & Fazio, G. Hormone and growth interactions of scions and size-controlling rootstocks of young apple trees. Plant Growth Regul. 78, 105–119 (2016).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Price, A. L., Jones, N. C. & Pevzner, P. A. De novo identification of repeat families in large genomes. Bioinformatics 21, i351–i358 (2005).
Bao, Z. & Eddy, S. R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12, 1269–1276 (2002).
Ellinghaus, D., Kurtz, S. & Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9, 18 (2008).
Ou, S. & Jiang, N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176, 1410–1422 (2018).
Bao, W., Kojima, K. K. & Kohany, O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015).
Zdobnov, E. M. & Apweiler, R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848 (2001).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Mendes, F. K., Vanderpool, D., Fulton, B. & Hahn, M. W. CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics 36, 5516–5518 (2020).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. GigaScience 10, giab008 (2021).
McKenna, A. et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Baum, B. R. PHYLIP: phylogeny inference package. Version 3.2. Joel Felsenstein. Q. Rev. Biol. 64, 539–541 (1989).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Zhang, C., Dong, S., Xu, J., He, W. & Yang, T. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 35, 1786–1788 (2019).
Marais, G. et al. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 14, e1005944 (2018).
Goel, M., Sun, H., Jiao, W. B. & Schneeberger, K. SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 20, 277 (2019).
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Fan, X. et al. Integrated multi-omics analysis uncovers roles of mdm-miR164b-MdORE1 in strigolactone-mediated inhibition of adventitious root formation in apple. Plant Cell Environ. 45, 3582–3603 (2022).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△Ct method. Methods 25, 402–408 (2001).
Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–W268 (2007).
Tang, H. et al. Synteny and collinearity in plant genomes. Science 320, 486–488 (2008).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Poplin, R. et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 36, 983–987 (2018).
Chu, C. simoncchu/RNAGlass: RNAGlass_v1.0.0 (v1.0.0). Zenodo https://doi.org/10.5281/zenodo.10146626 (2023).
Acknowledgements
This work was supported by the earmarked fund for the China Agricultural Research System (CARS-27 to Z.H.), 2115 Talent Development Program of China Agricultural University, the National Key Research and Development Program (2018YFD1000100 to W.L.), the National Natural Science Foundation of China (32172522 to Z.H. and 31801823 to W.L.) 111 Project (B17043) and the Growing Futures Fund from the New Zealand Institute for Plant and Food Research Limited.
Author information
Authors and Affiliations
Contributions
Z.H., W.L. and C.C. conceived and designed the project. W.L., Z.H., H.Z., H.L., Y.W. and H.S. collected and provided plant materials. W.L., C.C., H.L., H.S., C.H.D., S.W., Z.W., J.Y. and Y.X. assembled the genome and performed ASE analysis. C.C., W.L., H.L., C.H.D., H.S., Z.W. and Y.H. developed and validated the new pipeline for mobile mRNA identification. W.L., Yuqi Li and S.W. analyzed and interpreted population resequencing data. W.L., H.L., H.Z., Y.M., K.Z., B.Z., X.F., T.M.F. and E.L.-G. performed the validation and transformation experiments. Z.H., W.L., C.C., C.H.D., T.M.F., E.L.-G, H.L., H.Z., Y.W., H.S., S.W., Z.W. and Yuqi Li contributed to writing the paper. Yi Li and X.X. provided suggestions on data analysis and editing the paper. All authors read and approved the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Patrick Edger, Zhongchi Liu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–5, Methods, Figs. 1–24 and supporting data for Supplementary Fig. 21 (unprocessed gels).
Supplementary Tables
Supplementary Tables 1–32.
Source data
Source Data Fig. 4
Unprocessed gels for Fig. 4c.
Source Data Fig. 4
Statistical source data for Fig. 4g,i.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, W., Chu, C., Li, H. et al. Near-gapless and haplotype-resolved apple genomes provide insights into the genetic basis of rootstock-induced dwarfing. Nat Genet 56, 505–516 (2024). https://doi.org/10.1038/s41588-024-01657-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-024-01657-2