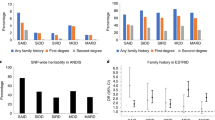
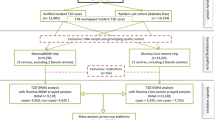
Abstract
Insulin resistance (IR) is a well-established risk factor for metabolic disease. The ratio of triglycerides to high-density lipoprotein cholesterol (TG:HDL-C) is a surrogate marker of IR. We conducted a genome-wide association study of the TG:HDL-C ratio in 402,398 Europeans within the UK Biobank. We identified 369 independent SNPs, of which 114 had a false discovery rate-adjusted P value < 0.05 in other genome-wide studies of IR making them high-confidence IR-associated loci. Seventy-two of these 114 loci have not been previously associated with IR. These 114 loci cluster into five groups upon phenome-wide analysis and are enriched for candidate genes important in insulin signaling, adipocyte physiology and protein metabolism. We created a polygenic-risk score from the high-confidence IR-associated loci using 51,550 European individuals in the Michigan Genomics Initiative. We identified associations with diabetes, hyperglyceridemia, hypertension, nonalcoholic fatty liver disease and ischemic heart disease. Collectively, this study provides insight into the genes, pathways, tissues and subtypes critical in IR.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
GWAS results from this study are available at the GWAS Catalog (study accessions GCST90295949–GCST90295954, all intervening numbers). UKBB genomic and phenotypic data supporting this publication are available upon application (https://ukbiobank.ac.uk). MGI individual-level data are not currently available to the public due to patient privacy requirements. Otherwise, all data used to generate figures can be found in supplementary tables, source data or in the above publicly available datasets. Source data are provided with this paper.
Code availability
The code to assign the gene labels is publicly available at https://doi.org/10.5281/zenodo.10182519 (ref. 89) and https://github.com/oliveriantonino/annotation_TGHDL.
References
Brown, A. E. & Walker, M. Genetics of insulin resistance and the metabolic syndrome. Curr. Cardiol. Rep. 18, 75 (2016).
Melvin, A., O’Rahilly, S. & Savage, D. B. Genetic syndromes of severe insulin resistance. Curr. Opin. Genet. Dev. 50, 60–67 (2018).
Mundi, M. S. et al. Evolution of NAFLD and its management. Nutr. Clin. Pract. 35, 72–84 (2020).
Ormazabal, V. et al. Association between insulin resistance and the development of cardiovascular disease. Cardiovasc. Diabetol. 17, 122 (2018).
Lee, J. M., Okumura, M. J., Davis, M. M., Herman, W. H. & Gurney, J. G. Prevalence and determinants of insulin resistance among U.S. adolescents: a population-based study. Diabetes Care 29, 2427–2432 (2006).
Ren, X. et al. Association between triglyceride to HDL-C ratio (TG/HDL-C) and insulin resistance in Chinese patients with newly diagnosed type 2 diabetes mellitus. PLoS ONE 11, e0154345 (2016).
Bonora, E. et al. Homeostasis model assessment closely mirrors the glucose clamp technique in the assessment of insulin sensitivity: studies in subjects with various degrees of glucose tolerance and insulin sensitivity. Diabetes Care 23, 57–63 (2000).
Stühlinger, M. C. et al. Relationship between insulin resistance and an endogenous nitric oxide synthase inhibitor. JAMA 287, 1420–1426 (2002).
Chen, J. Meta-Analysis of Glucose and Insulin-related Traits Consortium (MAGIC) et al. The trans-ancestral genomic architecture of glycemic traits. Nat. Genet. 53, 840–860 (2021).
Walford, G. A. et al. Genome-wide association study of the modified Stumvoll insulin sensitivity index identifies BCL2 and FAM19A2 as novel insulin sensitivity loci. Diabetes 65, 3200–3211 (2016).
Prokopenko, I. et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 10, e1004235 (2014).
Scott, R. A. et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat. Genet. 44, 991–1005 (2012).
Manning, A. K. et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 44, 659–669 (2012).
Dupuis, J. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 42, 105–116 (2010).
Iwani, N. A. et al. Triglyceride to HDL-C ratio is associated with insulin resistance in overweight and obese children. Sci. Rep. 7, 40055 (2017).
McLaughlin, T. et al. Use of metabolic markers to identify overweight individuals who are insulin resistant. Ann. Intern. Med. 139, 802–809 (2003).
Pantoja-Torres, B. et al. High triglycerides to HDL-cholesterol ratio is associated with insulin resistance in normal-weight healthy adults. Diabetes Metab. Syndr. 13, 382–388 (2019).
Chiang, J. K., Lai, N. S., Chang, J. K. & Koo, M. Predicting insulin resistance using the triglyceride-to-high-density lipoprotein cholesterol ratio in Taiwanese adults. Cardiovasc. Diabetol. 10, 93 (2011).
Gong, R. et al. Associations between TG/HDL ratio and insulin resistance in the US population: a cross-sectional study. Endocr. Connect. 10, 1502–1512 (2021).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Zhou, W. et al. Efficiently controlling for case–control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 50, 1335–1341 (2018).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Tang, J. et al. Obesity-associated family with sequence similarity 13, member A (FAM13A) is dispensable for adipose development and insulin sensitivity. Int. J. Obes. (Lond.) 43, 1269–1280 (2019).
Fathzadeh, M. et al. FAM13A affects body fat distribution and adipocyte function. Nat. Commun. 11, 1465 (2020).
Fernandes Silva, L., Vangipurapu, J., Kuulasmaa, T. & Laakso, M. An intronic variant in the GCKR gene is associated with multiple lipids. Sci. Rep. 9, 10240 (2019).
Li, X., Wang, F., Xu, M., Howles, P. & Tso, P. ApoA-IV improves insulin sensitivity and glucose uptake in mouse adipocytes via PI3K-Akt signaling. Sci. Rep. 7, 41289 (2017).
Nowak, M. et al. Insulin-mediated down-regulation of apolipoprotein A5 gene expression through the phosphatidylinositol 3-kinase pathway: role of upstream stimulatory factor. Mol. Cell. Biol. 25, 1537–1548 (2005).
Haas, M. E., Attie, A. D. & Biddinger, S. B. The regulation of ApoB metabolism by insulin. Trends Endocrinol. Metab. 24, 391–397 (2013).
Kim, J. Y., Tillison, K., Lee, J. H., Rearick, D. A. & Smas, C. M. The adipose tissue triglyceride lipase ATGL/PNPLA2 is downregulated by insulin and TNF-α in 3T3-L1 adipocytes and is a target for transactivation by PPARγ. Am. J. Physiol. Endocrinol. Metab. 291, E115–E127 (2006).
Knowles, J. W. et al. Identification and validation of N-acetyltransferase 2 as an insulin sensitivity gene. J. Clin. Investig. 125, 1739–1751 (2015).
Graham, S. E. et al. The power of genetic diversity in genome-wide association studies of lipids. Nature 600, 675–679 (2021).
Pulit, S. L. et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 28, 166–174 (2019).
Mahajan, A. et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 50, 1505–1513 (2018).
Chen, V. L. et al. Genome-wide association study of serum liver enzymes implicates diverse metabolic and liver pathology. Nat. Commun. 12, 816 (2021).
Hartiala, J. A. et al. Genome-wide analysis identifies novel susceptibility loci for myocardial infarction. Eur. Heart J. 42, 919–933 (2021).
Evangelou, E. et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 50, 1412–1425 (2018).
Stanzick, K. J. et al. Discovery and prioritization of variants and genes for kidney function in >1.2 million individuals. Nat. Commun. 12, 4350 (2021).
Chen, Y. et al. Genome-wide association meta-analysis identifies 17 loci associated with nonalcoholic fatty liver disease. Nat. Genet. 55, 1640–1650 (2023).
Day, F. et al. Large-scale genome-wide meta-analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. PLoS Genet. 14, e1007813 (2018).
Proud, C. G. Regulation of protein synthesis by insulin. Biochem. Soc. Trans. 34, 213–216 (2006).
Guillet, C., Masgrau, A., Walrand, S. & Boirie, Y. Impaired protein metabolism: interlinks between obesity, insulin resistance and inflammation. Obes. Rev. 13, 51–57 (2012).
Yang, Q. & Civelek, M. Transcription factor KLF14 and metabolic syndrome. Front. Cardiovasc. Med. 7, 91 (2020).
Teslovich, T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Shungin, D. et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 518, 187–196 (2015).
Small, K. S. et al. Regulatory variants at KLF14 influence type 2 diabetes risk via a female-specific effect on adipocyte size and body composition. Nat. Genet. 50, 572–580 (2018).
Mahdessian, H. et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc. Natl Acad. Sci. USA 111, 8913–8918 (2014).
Zabaneh, D. & Balding, D. J. A genome-wide association study of the metabolic syndrome in Indian Asian men. PLoS ONE 5, e11961 (2010).
Zhu, Y. et al. Susceptibility loci for metabolic syndrome and metabolic components identified in Han Chinese: a multi-stage genome-wide association study. J. Cell. Mol. Med. 21, 1106–1116 (2017).
Kristiansson, K. et al. Genome-wide screen for metabolic syndrome susceptibility loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ. Cardiovasc. Genet. 5, 242–249 (2012).
Kraja, A. T. et al. A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes 60, 1329–1339 (2011).
Agius, L., Chachra, S. S. & Ford, B. E. The protective role of the carbohydrate response element binding protein in the liver: the metabolite perspective. Front. Endocrinol. 11, 594041 (2020).
Abdul-Wahed, A., Guilmeau, S. & Postic, C. Sweet sixteenth for ChREBP: established roles and future goals. Cell Metab. 26, 324–341 (2017).
Ortega-Prieto, P. & Postic, C. Carbohydrate sensing through the transcription factor ChREBP. Front. Genet. 10, 472 (2019).
Arden, C. et al. Elevated glucose represses liver glucokinase and induces its regulatory protein to safeguard hepatic phosphate homeostasis. Diabetes 60, 3110–3120 (2011).
Lind, L. Genome-wide association study of the metabolic syndrome in UK Biobank. Metab. Syndr. Relat. Disord. 17, 505–511 (2019).
O’Donovan, G. et al. Fat distribution in men of different waist girth, fitness level and exercise habit. Int. J. Obes. (Lond.) 33, 1356–1362 (2009).
Paley, C.A. & Johnson, M. I. Abdominal obesity and metabolic syndrome: exercise as medicine? BMC Sports Sci. Med. Rehabil. 10, 7 (2018).
Shi, T. H., Wang, B. & Natarajan, S. The influence of metabolic syndrome in predicting mortality risk among US adults: importance of metabolic syndrome even in adults with normal weight. Prev. Chronic Dis. 17, E36 (2020).
Wang, K. et al. Differential roles of insulin like growth factor 1 receptor and insulin receptor during embryonic heart development. BMC Dev. Biol. 19, 5 (2019).
Holmes, D. I. & Zachary, I. The vascular endothelial growth factor (VEGF) family: angiogenic factors in health and disease. Genome Biol. 6, 209 (2005).
Kim, S., Ahn, C., Bong, N., Choe, S. & Lee, D. K. Biphasic effects of FGF2 on adipogenesis. PLoS ONE 10, e0120073 (2015).
Blázquez-Medela, A. M., Jumabay, M. & Boström, K. I. Beyond the bone: bone morphogenetic protein signaling in adipose tissue. Obes. Rev. 20, 648–658 (2019).
Yaghootkar, H. et al. Genetic evidence for a normal-weight ‘metabolically obese’ phenotype linking insulin resistance, hypertension, coronary artery disease, and type 2 diabetes. Diabetes 63, 4369–4377 (2014).
Bond, S. T., Calkin, A. C. & Drew, B. G. Sex differences in white adipose tissue expansion: emerging molecular mechanisms. Clin. Sci. (Lond.) 135, 2691–2708 (2021).
Brown, R. J. et al. The diagnosis and management of lipodystrophy syndromes: a multi-society practice guideline. J. Clin. Endocrinol. Metab. 101, 4500–4511 (2016).
Huang, Z., Xu, A. & Cheung, B. M. Y. The potential role of fibroblast growth factor 21 in lipid metabolism and hypertension. Curr. Hypertens. Rep. 19, 28 (2017).
Iizuka, K., Takao, K. & Yabe, D. ChREBP-mediated regulation of lipid metabolism: involvement of the gut microbiota, liver, and adipose tissue. Front. Endocrinol. 11, 587189 (2020).
Santoro, N. et al. Hepatic de novo lipogenesis in obese youth is modulated by a common variant in the GCKR gene. J. Clin. Endocrinol. Metab. 100, E1125–E1132 (2015).
Brouwers, M. C. G. J., Jacobs, C., Bast, A., Stehouwer, C. D. A. & Schaper, N. C. Modulation of glucokinase regulatory protein: a double-edged sword? Trends Mol. Med. 21, 583–594 (2015).
Chauhan, A., Singhal, A. & Goyal, P. TG/HDL ratio: a marker for insulin resistance and atherosclerosis in prediabetics or not? J. Fam. Med. Prim. Care 10, 3700–3705 (2021).
Cordero, A. & Alegria-Ezquerra, E. TG/HDL ratio as surrogate marker for insulin resistance. E J. Cardiol. Pract. 8, (2009).
Giannini, C. et al. The triglyceride-to-HDL cholesterol ratio: association with insulin resistance in obese youths of different ethnic backgrounds. Diabetes Care 34, 1869–1874 (2011).
Behiry, E. G., El Nady, N. M., AbdEl Haie, O. M., Mattar, M. K. & Magdy, A. Evaluation of TG-HDL ratio instead of HOMA ratio as insulin resistance marker in overweight and children with obesity. Endocr. Metab. Immune Disord. Drug Targets 19, 676–682 (2019).
Knight, M. G. et al. The TG/HDL-C ratio does not predict insulin resistance in overweight women of African descent: a study of South African, African American and West African women. Ethn. Dis. 21, 490–494 (2011).
Young, K. A. et al. The triglyceride to high-density lipoprotein cholesterol (TG/HDL-C) ratio as a predictor of insulin resistance, β-cell function, and diabetes in Hispanics and African Americans. J. Diabetes Complications 33, 118–122 (2019).
Maguire, L. H. et al. Genome-wide association analyses identify 39 new susceptibility loci for diverticular disease. Nat. Genet. 50, 1359–1365 (2018).
Dey, R., Schmidt, E. M., Abecasis, G. R. & Lee, S. A fast and accurate algorithm to test for binary phenotypes and its application to PheWAS. Am. J. Hum. Genet. 101, 37–49 (2017).
Zawistowski, M. et al. The Michigan Genomics Initiative: a biobank linking genotypes and electronic clinical records in Michigan Medicine patients. Cell Genom. 3, 100257 (2023).
Taliun, D. et al. LASER server: ancestry tracing with genotypes or sequence reads. Bioinformatics 33, 2056–2058 (2017).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Franzén, O. et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science 353, 827–830 (2016).
Machiela, M. J. & Chanock, S. J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557 (2015).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Oliveri, A. Code used to annotate the TG:HDL-C loci in the paper ‘comprehensive genetic study of the insulin resistance marker TG:HDL-C in the UK Biobank.’ Zenodo. https://doi.org/10.5281/zenodo.10182519 (2023).
Acknowledgements
The authors would like to acknowledge the MGI participants, Precision Health at the University of Michigan, the University of Michigan Medical School Central Biorepository and the University of Michigan Advanced Genomics Core for providing data and specimen storage, management, processing and distribution services, as well as the Center for Statistical Genetics in the Department of Biostatistics at the School of Public Health for genotype data curation, imputation and management in support of the research reported in this publication. A.O., A.K., A.P., X.D., Y.C., K.C.C., C.R., P.P., V.L.C., B.D.H. and E.K.S. supported in part by R01 DK106621 (to E.K.S.), R01 DK107904 (to E.K.S.), R01 DK128871 (to E.K.S.), R01 DK131787 (to E.K.S.) and/or the University of Michigan Department of Internal Medicine and/or The University of Michigan MBioFAR Award. R.J.R. is supported by F30 CA275039-02. H.B. is supported by F30 CA257292. Analyses in the UKBB were conducted under approved project 18120 (to E.K.S.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
E.K.S. contributed to the concept development, study design, data analysis and interpretation, writing of the manuscript and critical revision of the manuscript. A.O. and R.R. contributed to the study design, data analysis and interpretation, statistical analysis, drafting of the manuscript and critical review of the manuscript. X.D., Y.C. and C.R. contributed to data analysis and interpretation, statistical analysis and critical review of the manuscript. A.K., K.C.C., V.L.C., P.P., A.P., H.B. and B.H. contributed to data analysis and interpretation, drafting of the manuscript and critical review of the manuscript. All authors have critically reviewed the paper and approved the final version of this paper.
Corresponding author
Ethics declarations
Competing interests
V.L.C. received grant funding from KOWA and AstraZeneca. The Regents of the University of Michigan and E.K.S. have a pending patent on the use of systems and methods for analysis of samples associated with IR and related conditions. The rest of the authors do not have any conflicts of interest.
Peer review
Peer review information
Nature Genetics thanks Constantin Polychronakos and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note (abbreviations) and Supplementary Fig. 1.
Supplementary Tables
Supplementary Tables 1–25.
Source data
Source Data Fig. 2
Overlap of the 114 high-confidence IR-associated loci with insulin traits, novelty, gene annotation and expression of the gene in tissues.
Source Data Fig. 3
Heatmap and clusters of the 114 high-confidence IR-associated loci, and cluster PRSs and association in the UKBB.
Source Data Fig. 4
Results of the tissue, cell and system enrichment analysis.
Source Data Fig. 5
Results of the gene-sets enrichment analysis.
Source Data Fig. 6
Results of the PheWAS PRS analysis.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Oliveri, A., Rebernick, R.J., Kuppa, A. et al. Comprehensive genetic study of the insulin resistance marker TG:HDL-C in the UK Biobank. Nat Genet 56, 212–221 (2024). https://doi.org/10.1038/s41588-023-01625-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-023-01625-2