Abstract
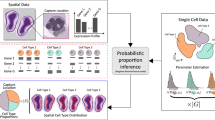
Spatial transcriptomics can reveal spatially resolved gene expression of diverse cells in complex tissues. However, the development of computational methods that can use the unique properties of spatial transcriptome data to unveil cell identities remains a challenge. Here we introduce SpiceMix, an interpretable method based on probabilistic, latent variable modeling for joint analysis of spatial information and gene expression from spatial transcriptome data. Both simulation and real data evaluations demonstrate that SpiceMix markedly improves on the inference of cell types and their spatial patterns compared with existing approaches. By applying to spatial transcriptome data of brain regions in human and mouse acquired by seqFISH+, STARmap and Visium, we show that SpiceMix can enhance the inference of complex cell identities, reveal interpretable spatial metagenes and uncover differentiation trajectories. SpiceMix is a generalizable analysis framework for spatial transcriptome data to investigate cell-type composition and spatial organization of cells in complex tissues.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The simulated data generated for this work are available at https://github.com/ma-compbio/SpiceMix. The spatial transcriptomic and single-cell datasets used in this study were obtained through publicly available repositories. The STARmap dataset is from https://www.starmapresources.org/data. The seqFISH+ dataset is from https://github.com/CaiGroup/seqFISH-PLUS. The Visium dataset is from https://research.libd.org/spatialLIBD, using the provided R commands. The snRNA-seq dataset of the human cortex is from https://portal.brain-map.org/atlases-and-data/rnaseq/human-multiple-cortical-areas-smart-seq. The scRNA-seq datasets of the mouse cortex are from the National Center for Biotechnology Information Gene Expression Omnibus (accession numbers GSE115746 and GSE71585).
Code availability
The source code of SpiceMix can be accessed at https://github.com/ma-compbio/SpiceMix and is downloadable from https://doi.org/10.5281/zenodo.725610759. For our comparisons against other methods, the following versions were used: Seurat v4.0.5, SpaGCN v1.0.0, BayesSpace v1.2.0, HMRF v1.3.3 and scHPF v0.5.0. The tool scDesign2 v0.1.0 for single-cell simulation was used as part of the process for generating the simulated data of approach II.
References
Arendt, D. et al. The origin and evolution of cell types. Nat. Rev. Genet. 17, 744–757 (2016).
Chen, X., Teichmann, S. A. & Meyer, K. B. From tissues to cell types and back: Single-cell gene expression analysis of tissue architecture. Ann. Rev. Biomed. Data Sci. 1, 29–51 (2018).
Consortium, H. et al. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019).
Lee, J. H. et al. Highly multiplexed subcellular RNA sequencing in situ. Science 343, 1360–1363 (2014).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Shah, S., Lubeck, E., Zhou, W. & Cai, L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 92, 342–357 (2016).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Moffitt, J. R. et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, eaau5324 (2018).
Eng, C.-H. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568, 235–239 (2019).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 341, eaat5691 (2018).
Rodriques, S. G. et al. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Vickovic, S. et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990 (2019).
Zhuang, X. Spatially resolved single-cell genomics and transcriptomics by imaging. Nat. Methods 18, 18–22 (2021).
Larsson, L., Frisén, J. & Lundeberg, J. Spatially resolved transcriptomics adds a new dimension to genomics. Nat. Methods 18, 15–18 (2021).
Lein, E., Borm, L. E. & Linnarsson, S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358, 64–69 (2017).
Palla, G., Fischer, D. S., Regev, A. & Theis, F. J. Spatial components of molecular tissue biology. Nat. Biotechnol. 40, 308–318 (2022).
Schapiro, D. et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods 14, 873–876 (2017).
Zhu, Q., Shah, S., Dries, R., Cai, L. & Yuan, G.-C. Identification of spatially associated subpopulations by combining scRNAseq and sequential fluorescence in situ hybridization data. Nat. Biotechnol. 36, 1183–1190 (2018).
Hu, J. et al. SpaGCN: Integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network. Nat. Methods 18, 1342–1351 (2021).
Jerby-Arnon, L. & Regev, A. Dialogue maps multicellular programs in tissue from single-cell or spatial transcriptomics data.Nat. Biotechnol. 40, 1467–1477 (2022).
Zhao, E. et al. Spatial transcriptomics at subspot resolution with bayesspace. Nat. Biotechnol. 39, 1375–1384 (2021).
Svensson, V., Teichmann, S. A. & Stegle, O. SpatialDE: identification of spatially variable genes. Nat. Methods 15, 343–346 (2018).
Arnol, D., Schapiro, D., Bodenmiller, B., Saez-Rodriguez, J. & Stegle, O. Modeling cell-cell interactions from spatial molecular data with spatial variance component analysis. Cell Rep. 29, 202–211 (2019).
Nitzan, M., Karaiskos, N., Friedman, N. & Rajewsky, N. Gene expression cartography. Nature 576, 132–137 (2019).
Sun, S., Zhu, J. & Zhou, X. Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies. Nat. Methods 17, 193–200 (2020).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Welch, J. D. et al. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177, 1873–1887 (2019).
Elosua-Bayes, M., Nieto, P., Mereu, E., Gut, I. & Heyn, H. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res 49, e50 (2021).
Biancalani, T. et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods 18, 1352–1362 (2021).
Lee, D. D. & Seung, H. S. Algorithms for non-negative matrixfactorization. Adv. Neural Inf. Process. Sys. 13, 556–562 (2000).
Maynard, K. R. et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci. 24, 425–436 (2021).
Sun, T., Song, D., Li, W. V. & Li, J. J. scDesign2: a transparent simulator that generates high-fidelity single-cell gene expression count data with gene correlations captured. Genome Biol. 22, 1–37 (2021).
Tasic, B. et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346 (2016).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Marques, S. et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329 (2016).
Zhao, C. et al. Dual regulatory switch through interactions of Tcf7l2/Tcf4 with stage-specific partners propels oligodendroglial maturation. Nat. Commun. 7, 10883 (2016).
Linington, C., Bradl, M., Lassmann, H., Brunner, C. & Vass, K. Augmentation of demyelination in rat acute allergic encephalomyelitis by circulating mouse monoclonal antibodies directed against a myelin/oligodendrocyte glycoprotein. Am. J. Pathol. 130, 443–454 (1988).
Tasic, B. et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018).
Zeisel, A. et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Marques, S. et al. Transcriptional convergence of oligodendrocyte lineage progenitors during development. Dev. Cell 46, 504–517 (2018).
Beiter, R. M. et al. Evidence for oligodendrocyte progenitor cell heterogeneity in the adult mouse brain. Sci. Rep. 12, 12921 (2022).
Levitin, H. M. et al. De novo gene signature identification from single-cell RNA-seq with hierarchical Poisson factorization. Mol. Syst. Biol. 15, e8557 (2019).
Allen Cell Types Database: Human Multiple Cortical Areas [Dataset] (Allen Institute for Brain Science, 2021); http://celltypes.brain-map.org/rnaseq
Zhang, M. et al. Spatially resolved cell atlas of the mouse primary motor cortex by merfish. Nature 598, 137–143 (2021).
Tan, S.-S. et al. Oligodendrocyte positioning in cerebral cortex is independent of projection neuron layering. Glia 57, 1024–1030 (2009).
Liu, Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183, 1665–1681 (2020).
Armingol, E., Officer, A., Harismendy, O. & Lewis, N. E. Deciphering cell–cell interactions and communication from gene expression. Nat. Rev. Genet. 22, 71–88 (2021).
Brunet, J.-P., Tamayo, P., Golub, T. R. & Mesirov, J. P. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl Acad. Sci. USA 101, 4164–4169 (2004).
Zhang, Y., Brady, M. & Smith, S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 20, 45–57 (2001).
Murphy, K. Machine Learning: A Probabilistic Perspective (MIT Press, 2012).
Besag, J. On the statistical analysis of dirty pictures. J. R. Stat. Soc. Ser. B 48, 259–279 (1986).
Gurobi Optimizer Reference Manual (Gurobi Optimization, 2020); http://www.gurobi.com
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. In Proc. 3rd International Conference on Learning Representations (ICLR, 2015).
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Pedregosa, F. et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Caliński, T. & Harabasz, J. A dendrite method for cluster analysis. Commun. Stat. Simul. Comput. 3, 1–27 (1974).
Gayoso, A., Shor, J., Carr, A. J., Sharma, R. & Pe’er, D. Doubletdetection (version v3.0) https://zenodo.org/record/6349517 (2020).
Chidester, B., Zhou, T., Alam, S. & Ma, J. SpiceMix (version v1.0.0) https://zenodo.org/record/7256107 (2022).
Acknowledgements
This work was supported in part by the National Institutes of Health Common Fund 4D Nucleome Program grant UM1HG011593 (J.M.), National Institutes of Health Common Fund Cellular Senescence Network Program grant UG3CA268202 (J.M.), National Institutes of Health grants R01HG007352 (J.M.) and R01HG012303 (J.M.), and National Science Foundation grant 1717205 (J.M.). J.M. is additionally supported by a Guggenheim Fellowship from the John Simon Guggenheim Memorial Foundation. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
Author information
Authors and Affiliations
Contributions
Conceptualization: B.C. and J.M. Methodology: B.C., T.Z. and J.M. Software: T.Z. and B.C. Investigation: B.C., T.Z., S.A. and J.M. Writing—original draft: B.C., T.Z. and J.M. Writing—review and editing: B.C., T.Z., S.A. and J.M. Funding acquisition: J.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Omer Bayraktar, Naveed Ishaque and Itai Yanai for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–23 and Note.
Supplementary Table 1
A table of the top 300 genes for each SpiceMix metagene for each dataset.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chidester, B., Zhou, T., Alam, S. et al. SpiceMix enables integrative single-cell spatial modeling of cell identity. Nat Genet 55, 78–88 (2023). https://doi.org/10.1038/s41588-022-01256-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-022-01256-z
This article is cited by
-
MENDER: fast and scalable tissue structure identification in spatial omics data
Nature Communications (2024)
-
Challenges and perspectives in computational deconvolution of genomics data
Nature Methods (2024)
-
Cellular neighborhood analysis in spatial omics reveals new tissue domains and cell subtypes
Nature Genetics (2024)
-
BANKSY unifies cell typing and tissue domain segmentation for scalable spatial omics data analysis
Nature Genetics (2024)
-
Representing and extracting knowledge from single-cell data
Biophysical Reviews (2024)