Abstract
Genome-wide association studies (GWAS) have identified many risk loci for Alzheimer’s disease (AD)1,2, but how these loci confer AD risk is unclear. Here, we aimed to identify loci that confer AD risk through their effects on brain protein abundance to provide new insights into AD pathogenesis. To that end, we integrated AD GWAS results with human brain proteomes to perform a proteome-wide association study (PWAS) of AD, followed by Mendelian randomization and colocalization analysis. We identified 11 genes that are consistent with being causal in AD, acting via their cis-regulated brain protein abundance. Nine replicated in a confirmation PWAS and eight represent new AD risk genes not identified before by AD GWAS. Furthermore, we demonstrated that our results were independent of APOE e4. Together, our findings provide new insights into AD pathogenesis and promising targets for further mechanistic and therapeutic studies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
Phenotypic, proteomic and transcriptomic data used in this manuscript are available via the AD Knowledge Portal (https://adknowledgeportal.org). The AD Knowledge Portal is a platform for accessing data, analyses and tools generated by the Accelerating Medicines Partnership (AMP-AD) Target Discovery Program and other National Institute on Aging (NIA)-supported programs to enable open-science practices and accelerate translational learning. The data, analyses and tools are shared early in the research cycle without a publication embargo on secondary use. Data are available for general research use according to the following requirements for data access and data attribution (https://adknowledgeportal.org/DataAccess/Instructions). Results of the pQTL analysis, protein weights and transcript weights described in this manuscript can be accessed at https://doi.org/10.7303/syn23627957.
References
Jansen, I. E. et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019).
Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 (2019).
Ballard, C. et al. Alzheimer’s disease. Lancet 377, 1019–1031 (2011).
Wingo, A. P. et al. Shared proteomic effects of cerebral atherosclerosis and Alzheimer’s disease on the human brain. Nat. Neurosci. 23, 696–700 (2020).
Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016).
Beach, T. G. et al. Arizona Study of Aging and Neurodegenerative Disorders and Brain and Body Donation Program. Neuropathology 35, 354–389 (2015).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Nicolas, A. et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 97, 1268–1283.e6 (2018).
Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019).
Nagel, M. et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat. Genet. 50, 920–927 (2018).
Yengo, L. et al. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 27, 3641–3649 (2018).
Pulit, S. L. et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 28, 166–174 (2019).
Wu, Y. et al. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat. Commun. 9, 918 (2018).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Wainberg, M. et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 51, 592–599 (2019).
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Gusev, A. et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 50, 538–548 (2018).
Huckins, L. M. et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat. Genet. 51, 659–674 (2019).
Raj, T. et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat. Genet. 50, 1584–1592 (2018).
Bennett, D. A. et al. Religious Orders Study and Rush Memory and Aging Project. J. Alzheimers Dis. 64, S161–S189 (2018).
Mertins, P. et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography–mass spectrometry. Nat. Protoc. 13, 1632–1661 (2018).
De Jager, P. L. et al. A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci. Data 5, 180142 (2018).
Purcell, S. et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 81, 559–575 (2007).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Abecasis, G. R. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Allen, M. et al. Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci. Data 3, 160089 (2016).
Wang, M. et al. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci. Data 5, 180185 (2018).
Wan, Y. W. et al. Meta-analysis of the alzheimer’s disease human brain transcriptome and functional dissection in mouse models. Cell Rep. 32, 107908 (2020).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Freedman, M. L. et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 36, 388–393 (2004).
Wu, L. et al. A transcriptome-wide association study of 229,000 women identifies new candidate susceptibility genes for breast cancer. Nat. Genet. 50, 968–978 (2018).
Sieberts, S. K. et al. Large eQTL meta-analysis reveals differing patterns between cerebral cortical and cerebellar brain regions. Sci. Data 7, 340 (2020).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Acknowledgements
We thank the participants of the ROS, MAP, Mayo, Mount Sinai Brain Bank and Banner Sun Health Research Institute Brain and Body Donation Program for their time and participation. The following National Institutes of Health (NIH) grants supported this work: P30 AG066511 (A.I.L.); P30 AG10161 (D.A.B.); P30 NS055077 (A.I.L.); P50 AG025688 (A.I.L.); R01 AG015819 (D.A.B.); R01 AG017917 (D.A.B.); R01 AG053960 (N.T.S.); R01 AG056533 (T.S.W., A.P.W.); R01 AG057911 (N.T.S.); R01 AG061800 (N.T.S.); R56 AG060757 (T.S.W.); R56 AG062256 (T.S.W.); RC2 AG036547 (D.A.B.); RF1 AG057470 (T.S.W.); U01 AG046152 (P.L.D.); U01 AG046161 (A.I.L.); U01 AG061356 (P.L.D.); U01 AG061357 (A.I.L.); and U01 MH115484 (A.P.W.). NIH grants include those that supported the Accelerating Medicine Partnership for AD, the National Institute of Neurological Disorders and Stroke Emory Neuroscience Core and Goizueta Alzheimer’s Disease Research Center (ADRC) at Emory University, the Rush University ADRC and Arizona State University ADRC that made this work possible. The following Veterans Administration grants supported this work: I01 BX003853 (A.P.W.) and IK4 BX005219 (A.P.W.). The Brain and Body Donation Program has been supported by the NIH, the Arizona Department of Health Services, the Arizona Biomedical Research Commission and the Michael J. Fox Foundation for Parkinson’s Research. Additional support includes grants from the Alzheimer’s Association (N.T.S.), Alzheimer’s Research UK (N.T.S.), the Michael J. Fox Foundation for Parkinson’s Research (N.T.S.) and the Weston Brain Institute Biomarkers Across Neurodegenerative Diseases Grant 11060 (N.T.S.). The views expressed in this work do not necessarily represent the views of the Veterans Administration or the United States Government.
Author information
Authors and Affiliations
Contributions
A.P.W. and T.S.W. conceptualized and designed the study. A.P.W., D.M.D., E.B.D., T.G.B., E.M.R., P.L.D., J.J.L., D.A.B., N.T.S., A.I.L. and T.S.W. acquired the data. A.P.W., Y.L., E.S.G., J.G., B.A.L. and T.S.W. conducted the analyses. A.P.W., Y.L., E.S.G., J.G., B.A.L., E.B.D., C.R., M.P.E., J.J.L., D.A.B., N.T.S., A.I.L. and T.S.W. interpreted the data. A.P.W. and T.S.W. wrote the first draft of the manuscript. All authors critically revised and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Genetics thanks Towfique Raj and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
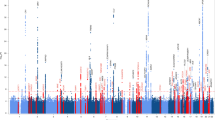
Extended Data Fig. 1 Quantile-quantile plots for the discovery and replication PWAS of AD.
Quantile-quantile plot for a, the discovery PWAS of AD (λ = 1.36; λ1000 = 1.003) and b, confirmatory PWAS of AD (λ = 1.39; λ1000 = 1.003).
Extended Data Fig. 2 Overlap of significant genes between AD and other traits.
Overlap between results of the AD PWAS and PWAS for other traits. All the PWAS used the discovery ROS/MAP proteomic dataset (n = 376) and GWAS summary results from Caucasian individuals. The following outcomes were tested: clinical AD GWAS (N = 63,926), amyotrophic lateral sclerosis (ALS; N = 80,610), body mass index (BMI; N = 681,275), height (N = 693,529), neuroticism (N = 390,278), Parkinson’s disease (PD; N = 1,474,097), and waist-to-hip ratio adjusting for BMI (WHRadjBMI; N = 694,649). Significant genes considered for overlap are those with FDR p < 0.05.
Extended Data Fig. 3 Quantile-quantile plot for the TWAS of AD.
Quantile-quantile plot for the TWAS of AD (λ = 1.22; λ1000 = 1.002).
Extended Data Fig. 4 Single cell-type expression.
Single-cell type expression for AD PWAS-significant genes with evidence of causality in AD. Using human brain single-cell RNA-sequencing data profiled from the dPFC, we found that 6 genes (of the 11 genes) had evidence of enrichment in a cell type at FDR p < 0.05. Enrichment testing was performed using Wilcoxon rank sum test, as implemented by the Seurat package, and multiple testing was accounted for by FDR adjusted for 17,775 tested genes. CARHSP1 showed enrichment in oligodendrocytes. CTSH showed enrichment in astrocytes and microglia. DOC2A, ICA1L, PLEKHA1, and SNX32 were enriched in excitatory neurons.
Extended Data Fig. 5 Genetic principal components of genetic ancestry for each dataset.
Genetic principal components of genetic ancestry for each dataset. The first two genetic principal components for individuals in each dataset are plotted (grey boxes) with individuals from the 1000 Genomes CEU dataset (purple triangles) for a, the discovery proteomic dataset, b, the replication proteomic dataset, and c, the transcriptomic dataset.
Supplementary information
Supplementary Information
List of supplementary tables
Supplementary Tables
Supplementary Tables 1–20
Source data
Source Data Extended Data Fig. 4
Statistical source data
Source Data Extended Data Fig. 5
Statistical source data
Rights and permissions
About this article
Cite this article
Wingo, A.P., Liu, Y., Gerasimov, E.S. et al. Integrating human brain proteomes with genome-wide association data implicates new proteins in Alzheimer’s disease pathogenesis. Nat Genet 53, 143–146 (2021). https://doi.org/10.1038/s41588-020-00773-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-020-00773-z
This article is cited by
-
Associations between genetically predicted plasma protein levels and Alzheimer’s disease risk: a study using genetic prediction models
Alzheimer's Research & Therapy (2024)
-
Decreased CNNM2 expression in prefrontal cortex affects sensorimotor gating function, cognition, dendritic spine morphogenesis and risk of schizophrenia
Neuropsychopharmacology (2024)
-
Identifying novel proteins for suicide attempt by integrating proteomes from brain and blood with genome-wide association data
Neuropsychopharmacology (2024)
-
TOPMed imputed genomics enhances genomic atlas of the human proteome in brain, cerebrospinal fluid, and plasma
Scientific Data (2024)
-
Genome-wide association study implicates lipid pathway dysfunction in antipsychotic-induced weight gain: multi-ancestry validation
Molecular Psychiatry (2024)