Abstract
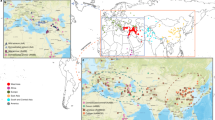
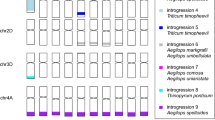
Introgression is a potential source of beneficial genetic diversity. The contribution of introgression to adaptive evolution and improvement of wheat as it was disseminated worldwide remains unknown. We used targeted re-sequencing of 890 diverse accessions of hexaploid and tetraploid wheat to identify wild-relative introgression. Introgression, and selection for improvement and environmental adaptation, each reduced deleterious allele burden. Introgression increased diversity genome wide and in regions harboring major agronomic genes, and contributed alleles explaining a substantial proportion of phenotypic variation. These results suggest that historic gene flow from wild relatives made a substantial contribution to the adaptive diversity of modern bread wheat.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data have been deposited in the European Variation Archive (EVA) under project PRJEB31218 and NCBI SRA under project PRJNA517692, and are available for viewing and download from http://wheatgenomics.plantpath.ksu.edu/1000EC.
Change history
13 June 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Nesbitt, M. & Samuel, D. From staple crop to extinction? The archaeology and history of the hulled wheats. in Proc. 1st Int. Workshop Hulled Wheats (eds Padulosi, S. et al.) 41–100 (Italy International Plant Genetic Resources Institute, 1996).
Tanno, K.-I. & Willcox, G. How fast was wild wheat domesticated? Science 311, 1886 (2006).
Luo, M.-C. et al. The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor. Appl. Genet. 114, 947–959 (2007).
Ozkan, H., Willcox, G., Graner, A., Salamini, F. & Kilian, B. Geographic distribution and domestication of wild emmer wheat (Triticum dicoccoides). Genet. Resour. Crop Evol. 58, 11–53 (2011).
Kihara, H. Discovery of the DD-analyser, one of the ancestors of Triticum vulgare. Agric. Hortic. 19, 889–890 (1944).
Dvorak, J., Luo, M. C., Yang, Z. L. & Zhang, H. B. The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 97, 657–670 (1998).
Smith, O. et al. Sedimentary DNA from a submerged site reveals wheat in the British Isles 8000 years ago. Science 347, 998–1001 (2014).
Long, T. et al. The early history of wheat in China from 14C dating and Bayesian chronological modelling. Nat. Plants 4, 272–279 (2018).
Haudry, a et al. Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol. Biol. Evol. 24, 1506–1517 (2007).
Akhunov, E. D. et al. Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genomics 11, 702 (2010).
Kerber, E. R. Wheat: reconstitution of the tetraploid component (AABB) of hexaploids. Science 143, 253–255 (1964).
Dvorak, J., Luo, M. & Akhunov, E. D. N. I. Vavilov' s theory of centres of diversity in the light of current understanding of wheat diversity, domestication and evolution. Czech. J. Genet. Plant Breed. 47, 1–8 (2011).
Dvorak, J., Akhunov, E. D., Akhunov, A. R., Deal, K. R. & Luo, M.-C. Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol. Biol. Evol. 23, 1386–1396 (2006).
Salojärvi, J. et al. Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat. Genet. 49, 904–912 (2017).
Rendón-anaya, M. et al. Genomic history of the origin and domestication of common bean unveils its closest sister species. Genome Biol. 18, 1–17 (2017).
Wang, L. et al. The interplay of demography and selection during maize domestication and expansion. Genome Biol. 18, 1–13 (2017).
Hardigan, M. A. et al. Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato. Proc. Natl Acad. Sci. USA 114, E9999–E10008 (2017).
Hübner, S. et al. Islands and streams: clusters and gene flow in wild barley populations from the Levant. Mol. Ecol. 21, 1115–1129 (2012).
International Wheat Genome Sequencing Consortium. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191 (2018).
Jordan, K. et al. A haplotype map of allohexaploid wheat reveals distinct patterns of selection on homoeologous genomes. Genome Biol. 16, 48 (2015).
Liu, Q., Zhou, Y., Morrell, P. L., Gaut, B. S. & Ge, S. Deleterious variants in Asian rice and the potential cost of domestication. Mol. Biol. Evol. 34, 908–924 (2017).
Mezmouk, S. & Ross-Ibarra, J. The pattern and distribution of deleterious mutations in maize. G3 (Bethesda) 4, 163–171 (2014).
Avni, R. et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 97, 93–97 (2017).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Cavanagh, C. R. et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl Acad. Sci. USA 110, 8057–8062 (2013).
Wang, S. et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796 (2014).
Poets, A. M. et al. The effects of both recent and long-term selection and genetic drift are readily evident in North American barley breeding populations. G3 (Bethesda) 6, 609–622 (2016).
Patterson, N. et al. Ancient admixture in human history. Genetics 192, 1065–1093 (2012).
Martin, S. H., Davey, J. W. & Jiggins, C. D. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol. Biol. Evol. 32, 244–257 (2015).
Smith, J. & Kronforst, M. R. Do Heliconius butterfly species exchange mimicry alleles? Biol. Lett. 9, 1–4 (2013).
Hufford, M. B. et al. The genomic signature of crop-wild introgression in maize. PLoS Genet. 9, e1003477 (2013).
Nave, M., Avni, R., Ben-Zvi, B., Hale, I. & Distelfeld, A. QTLs for uniform grain dimensions and germination selected during wheat domestication are co-located on chromosome 4B. Theor. Appl. Genet. 129, 1303–1315 (2016).
Simons, K. J. et al. Molecular characterization of the major wheat domestication gene Q. Genetics 172, 547–555 (2006).
Günther, T. & Coop, G. Robust identification of local adaptation from allele frequencies. Genetics 195, 205–220 (2013).
Chen, H., Patterson, N. & Reich, D. Population differentiation as a test for selective sweeps. Genome Res. 20, 393–402 (2010).
Kant, S., Bi, Y. & Rothstein, S. J. Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 62, 1499–1509 (2011).
Forde, B. G. Glutamate signalling in roots. J. Exp. Bot. 65, 779–787 (2014).
Lu, G. et al. Application of T-DNA activation tagging to identify glutamate receptor-like genes that enhance drought tolerance in plants. Plant Cell Rep. 33, 617–631 (2014).
Kiba, T., Krapp, A. & Science, R. Plant nitrogen acquisition under low availability: regulation of uptake and root architecture. Plant Cell Physiol. 57, 707–714 (2016).
Kono, T. J. Y. et al. The role of deleterious substitutions in crop genomes. Mol. Biol. Evol. 33, 2307–2317 (2016).
Jordan, K. W. et al. The genetic architecture of genome‐wide recombination rate variation in allopolyploid wheat revealed by nested association mapping. Plant J. 95, 1039–1054 (2018).
Kilian, B. et al. Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Mol. Biol. Evol. 24, 217–227 (2007).
Choulet, F. et al. Structural and functional partitioning of bread wheat chromosome 3B. Science 345, 1249721 (2014).
Akhunova, A. R., Matniyazov, R. T., Liang, H. & Akhunov, E. D. Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Genomics 11, 1–16 (2010).
Veitia, R. A., Bottani, S. & Birchler, J. A. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 24, 390–397 (2008).
Gusev, A. et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 95, 535–552 (2014).
Peleg, Z., Fahima, T., Korol, A. B., Abbo, S. & Saranga, Y. Genetic analysis of wheat domestication and evolution under domestication. J. Exp. Bot. 62, 5051–5061 (2011).
Stitzer, M. C. & Ross-Ibarra, J. Maize domestication and gene interaction. New Phytol. 220, 395–408 (2018).
Morrell, P. L., Buckler, E. S. & Ross-Ibarra, J. Crop genomics: advances and applications. Nat. Rev. Genet. 13, 85–96 (2011).
Krasileva, K. V. et al. Uncovering hidden variation in polyploid wheat. Proc. Natl Acad. Sci. USA 114, 913–E921 (2017).
Ramu, P. et al. Cassava haplotype map highlights fixation of deleterious mutations during clonal propagation. Nat. Genet. 49, 959–963 (2017).
Zhao, Y. et al. Genome-based establishment of a high-yielding heterotic pattern for hybrid wheat breeding. Proc. Natl Acad. Sci. USA 112, 15624–15629 (2015).
Hao, Y. et al. Patterns of population variation in two paleopolyploid eudicot lineages suggest that dosage-based selection on homeologs is long-lived. Genome Biol. Evol. 10, 999–1011 (2018).
Yu, X., Woolliams, J. A. & Meuwissen, T. H. E. Prioritizing animals for dense genotyping in order to impute missing genotypes of sparsely genotyped animals. Genet. Sel. Evol. 46, 1–8 (2014).
Browning, B. L. & Browning, S. R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194, 459–471 (2013).
Thuillet, A.-C., Bataillon, T., Poirier, S., Santoni, S. & David, J. L. Estimation of long-term effective population sizes through the history of durum wheat using microsatellite data. Genetics 169, 1589–1599 (2005).
Keightley, P. D. & Jackson, B. C. Inferring the probability of the derived vs. the ancestral allelic state at a polymorphic site. Genetics 209, 897–906 (2018).
Luo, M.-C. et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551, 498–502 (2017).
Ling, H.-Q. et al. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496, 87–90 (2013).
Mascher, M. et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433 (2017).
De Baets, G. et al. SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 40, D935–D939 (2012).
Jakobsson, M. & Rosenberg, N. A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806 (2007).
Green, R. E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722 (2010).
Neph, S. et al. BEDOPS: high-performance genomic feature operations. Bioinformatics 28, 1919–1920 (2012).
Conesa, A. & Götz, S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 619832 (2008).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 57, 289–300 (1995).
Supek, F., Bošnjak, M., Škunca, N. & Šmuc, T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 (2011).
Fischer, R. A. & Maurer, R. Drought resistance in spring wheat cultivars: I. Grain yield responses. Aust. J. Agric. Res 29, 897–912 (1978).
Yang, J. et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 43, 519–525 (2011).
Acknowledgements
This project was supported by the Agriculture and Food Research Initiative Competitive Grants 2017-67007-25939 (Wheat-CAP) and 2016-67013-24473 from the USDA National Institute of Food and Agriculture, and grants from the Bill and Melinda Gates Foundation and Kansas Wheat Commission. Exome sequencing of Canadian wheat cultivars was supported through the Canadian Triticum Applied Genomics grant funded by Genome Canada, Genome Prairie, Saskatchewan Ministry of Agriculture, and the Western Grains Research Foundation. P.L.M. was supported by grant IOS-1339393 from the US National Science Foundation. Corteva Agriscience, Agriculture Division of DowDuPont provided financial support through collaboration with Agriculture Victoria Services enabling the development of the SNP dataset and technologies used in this manuscript. The authors would like to thank International Wheat Genome Sequencing Consortium for providing access to wheat genome sequence under Toronto agreement, D. Andresen for assistance with the computing resources of the KSU Beocat cluster funded by NSF grant ACI-144054 and K. Jordan for valuable suggestions and editing the manuscript.
Author information
Authors and Affiliations
Contributions
F.H. led the bioinformatic and statistical analyses of data and helped to draft the first version of the manuscript. R.P. led phenotypic analyses. F.S. contributed to genomic analyses. S.K. was responsible for field trials and phenotype collection. G.K.-G. contributed to bioinformatic analyses of data. P.K. was responsible for exome sequencing of most wheat lines. K.F. was responsible for exome sequencing and 90K SNP data analyses. A.F. contributed to generating wild emmer exome capture data. P.H., K.W., R.K., R.C. and C.P. generated and contributed exome sequencing data for wild and domesticated emmer, and Canadian wheat cultivars. A.A. contributed to exome capture of wild and domesticated emmer, and wheat. P.L.M. contributed to data interpretation and manuscript writing. C.P., J.P.D., S.R.W. and G.S. contributed to project design. B.H., H.D. and J.T. contributed to project coordination and data analyses. M.H. provided project leadership, coordinated data collection and next generation sequencing (NGS) data analyses, and contributed to manuscript writing. E.A. conceived the idea, coordinated data collection and NGS data analyses and data interpretation, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–13, Supplementary Tables 2, 3, 14–16, 19 and 20, and Supplementary Note
Supplementary Table 1
List of hexaploid wheat accessions used in the study.
Supplementary Table 4
Genetic differentiation between wheat landraces and cultivars
Supplementary Table 5
Distribution of population-based fd statistic and frequency of introgression (FI) across genome.
Supplementary Table 6
Distribution of introgression statistics across the wheat genome
Supplementary Table 7
Ancestral allelic states inferred using multiple outgroup species
Supplementary Table 8
Locations of introgressed genomic regions (IGRs).
Supplementary Table 9
Climatic and bioclimatic data from WorldClim database used in Bayenv analyses
Supplementary Table 10
Genomic regions associated with environmental adaptation
Supplementary Table 11
The genomic regions showing the evidence of improvement selection
Supplementary Table 12
The genomic regions shared by all three scans for introgression, XP-CLR and Bayenv
Supplementary Table 13
GO terms enriched for genes located in the regions detected using the XP-CLR, Bayenv and fd – statistics analyses
Supplementary Table 17
Overlap of GWAS signals with introgression
Supplementary Table 18
Homoeolog-specific bias in gene expression between introgressed (I) and non-introgressed (NI) genomic regions
Rights and permissions
About this article
Cite this article
He, F., Pasam, R., Shi, F. et al. Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat Genet 51, 896–904 (2019). https://doi.org/10.1038/s41588-019-0382-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0382-2
This article is cited by
-
Introgressions lead to reference bias in wheat RNA-seq analysis
BMC Biology (2024)
-
Significant genomic introgression from grey junglefowl (Gallus sonneratii) to domestic chickens (Gallus gallus domesticus)
Journal of Animal Science and Biotechnology (2024)
-
Lifting of the 1,000 wheat exome project SNPs from Triticum aestivum cv. Chinese Spring assembly RefSeq v1.0 to RefSeq v2.1
BMC Research Notes (2023)
-
Genetic diversity and population structure of modern wheat (Triticum aestivum L.) cultivars in Henan Province of China based on SNP markers
BMC Plant Biology (2023)