Abstract
Maize is an important crop with a high level of genome diversity and heterosis. The genome sequence of a typical female line, B73, was previously released. Here, we report a de novo genome assembly of a corresponding male representative line, Mo17. More than 96.4% of the 2,183 Mb assembled genome can be accounted for by 362 scaffolds in ten pseudochromosomes with 38,620 annotated protein-coding genes. Comparative analysis revealed large gene-order and gene structural variations: approximately 10% of the annotated genes were mutually nonsyntenic, and more than 20% of the predicted genes had either large-effect mutations or large structural variations, which might cause considerable protein divergence between the two inbred lines. Our study provides a high-quality reference-genome sequence of an important maize germplasm, and the intraspecific gene order and gene structural variations identified should have implications for heterosis and genome evolution.
Similar content being viewed by others
Main
Maize (Zea mays) is a classical genetic model and an important crop worldwide. The maize genome exhibits high levels of genetic diversity among different inbred lines1,2,3,4,5,6. Owing to the high level of intraspecific genome diversity, hybrid maize resulting from crosses between lines from different heterotic groups shows extremely high levels of hybrid vigor. Consequently, the adoption of hybrid maize has grown rapidly since its introduction nearly a century ago7. Currently, most modern maize varieties are hybrids. Reid yellow dent (represented by inbred B73) and Lancaster (represented by inbred Mo17) are the two best-known maize variety groups. The hybrid generated by crossing B73 with Mo17 had long been the most commonly grown hybrid in America and other countries8, and the derived materials are still widely used in many maize breeding programs. B73/Mo17 is therefore the most common pair of maize inbred lines in many genetic and molecular studies, such as map-based cloning9 or exploring the molecular basis of heterosis10 and genetic imprinting11,12,13. The intermated population of B73 and Mo17 is the most prominent maize genetic-mapping population14. The release of the draft genome assembly of inbred line B73 in 2009 (ref. 6) was an important milestone in the maize community15. The draft genome sequences of the maize PH207 inbred line assembled from short reads have also been reported16. The precise genomic arrangement of maize has recently been demonstrated to be resolvable through assembly of PacBio long reads17, which are up to 40–60 kb long, albeit with a relatively high error rate18. Long-read assembly has been used to generate several other plant and animal genomes19,20,21,22. Recently, a high-quality whole-genome assembly of B73 (RefGen_v4) has been generated through single-molecule technologies and the BioNano Irys system23, thus resulting in major improvements relative to the earlier assembly.
Here, we report the assembly of a high-quality Mo17 reference genome through single-molecule sequencing and BioNano optical-mapping technologies. The generation of an additional reference genome provides an unprecedented opportunity for extensive comparison of intraspecific genome diversity in maize. By aligning the B73 and Mo17 genomes, we found 9,867,466 SNPs; 1,422,446 small insertions/deletions (indels, length shorter than 100 bp); and more than 25 MB of presence/absence-variation (PAV, length longer than 500 bp) sequences between the two representative maize genomes. Notably, our comparative genomics analysis uncovered extensive intraspecific gene-order variation: approximately 10% of genes were mutually nonsyntenic between B73 and Mo17. In addition, more than 20% of the annotated genes had large-effect mutations or large structural variations in B73 compared with Mo17. These large gene-order and gene structural variations were also observed in a comparison of the PH207 genome with the B73 and Mo17 genomes.
Results
Genome sequencing and assembly
We sequenced and assembled the genome of Mo17 through a combination of three technologies: single-molecule real time (SMRT) sequencing with the PacBio Sequel platform, paired-end sequencing with the Illumina HiSeq platform and optical genome mapping with the BioNano Genomics Irys System (Methods). The initial assembly of 24.11 million PacBio long reads (200.8 Gb in total), representing ~90× sequencing coverage of the Mo17 genome, resulted in a 2,148-Mb assembly with a contig N50 size of 1.48 Mb (Table 1 and Supplementary Tables 1 and 2). The assembled contigs were polished with Pilon24 with 251.8-Gb high-quality paired-end PCR-free reads, then scaffolded by optical maps assembled with 267.7-Gb BioNano molecules. The final assembly contains 2,560 scaffolds with a scaffold N50 size of 10.2 Mb (Table 1 and Supplementary Table 2). The total assembly size of the Mo17 genome is 2,183 Mb, a genome size similar to that of the recently updated B73 genome (2,106 Mb)23. Approximately 97.2% (1,399 of 1,440) of embryophyta genes were detected in our assembly according to BUSCO25, a percentage similar to that for the B73 genome (97.3%), thus indicating the near completeness of our assembly. To evaluate the quality of the assembled Mo17 genome, we downloaded nine previously published Mo17 bacterial artificial chromosome (BAC) sequences from GenBank and aligned them to the scaffolds with BLAT26. All nine BAC sequences were covered by a single scaffold with high consistency and coverage (Supplementary Table 3), thus indicating high quality of the assembled genome. With a high-density genetic map containing approximately 4.4 million genotype-by-sequencing (GBS) tags27, we anchored and oriented 362 scaffolds onto ten pseudochromosomes, which account for 96.42% (2,104 Mb) of the genome assembly (Supplementary Table 4). The alignment of the Mo17 and B73 genomes with the anchored GBS tags27 showed high consistency with respect to the position and orientation of the anchored scaffolds (Supplementary Fig. 1).
Genome annotation
Repetitive elements, major components of complex genomes, are widely dispersed throughout the genome and have multiple roles in driving genome evolution28. In total, approximately 83.83% of the Mo17 assembly sequences were annotated as repetitive elements, including retrotransposons (75.24%), DNA transposons (6.12%) and unclassified elements (1.72%) (Supplementary Table 5). The families of Gypsy and Copia retrotransposons represented approximately 48.63% and 25.57% of the Mo17 assembly sequences, respectively. The composition of the different classes of repetitive DNA in Mo17 was highly similar to that in the B73 and PH207 genomes (Supplementary Table 5).
To examine transposon activity, we identified a total of 73,459, 74,160 and 50,402 high-confidence full-length long terminal repeat (LTR) retrotransposons in the B73, Mo17 and PH207 genomes, respectively. The expansion of LTR retrotransposons in maize occurred mainly within the past 1 million years in both the B73 and Mo17 genomes (Supplementary Fig. 2), in agreement with previous estimates based on analysis of selected regions of the maize genome29,30. A relatively lower percentage of young LTR retrotransposons was seen in the PH207 genome assembly, because highly similar copies were collapsed when assembled from short reads (Supplementary Fig. 2). Compared with the older LTR retrotransposons, which were more abundant in pericentromeric regions, the younger LTR retrotransposons were enriched in euchromatic regions (Supplementary Fig. 3), in agreement with findings from previous studies31,32.
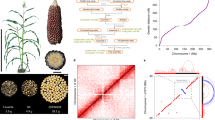
To annotate the protein-coding genes in the Mo17 genome, we combined results obtained from protein-homology-based prediction, RNA-seq-based prediction and ab initio prediction (Methods), an approach similar to that used for the annotation of the B73 (refs 23,33) and PH207 (ref. 16) genomes. In total, 38,620 high-confidence protein-coding genes were predicted in the Mo17 genome, and 55% of the exons of the predicted genes were supported by RNA-seq data from five different tissues with at least 90% coverage (Supplementary Tables 6 and 7). In total, 37,830 (97.95%) of the Mo17 predicted genes were allocated in ten pseudochromosomes (Supplementary Table 4). Protein-coding genes were primarily within chromosome arms and were inversely correlated with transposable-element density (Fig. 1).
a, Distribution of subgenome 1 (red) and subgenome 2 (yellow) in the B73 and Mo17 genomes. Gray indicates regions without assigned subgenome information. b,c, Transposable-element (b) and gene (c) density in sliding windows of 1Mb. d,e, numbers of SNPs (d) and indels (e) in sliding windows of 1Mb. f, Distribution of PAV sequences. g, Gene pairs between B73 and Mo17 genomes identified with the best-hit method.
Global genome comparison of B73, Mo17 and PH207
When the pseudochromosomes of Mo17 were aligned to the pseudochromosomes of B73 (ref. 34), approximately 61.18% of the Mo17 genome sequence matched in one-to-one syntenic blocks with 61.13% of the B73 genome sequence (Supplementary Fig. 4 and Supplementary Table 8). The genome-wide proportion of the regions that nearly matched between B73 and Mo17 was slightly higher than that estimated from a previous analysis of sequenced BAC clones of four selected regions35. The nonsyntenic sequences between the two genomes were mostly transposable elements, and the remainder comprised dispersed genes and inbred line-specific low-copy sequences. Similarly, we found 1,071.7 Mb (50.93%) of the Mo17 genome sequence and 1,101.7 Mb (52.3%) of the B73 genome sequence matching in syntenic blocks with 1,071.6 Mb (52.01%) and 1,101.4 Mb (53.46%) of the current PH207 genome sequences, respectively (Supplementary Table 8).
By comparing the two genomes, we identified 12,936 B73-specific genomic segments (12.96 Mb in total) and 12,939 Mo17-specific genomic segments (12.2 Mb in total) longer than 500 bp. Most (98.7%) of these PAV sequences were shorter than 5 kb (Supplementary Fig. 5). We found 200 and 126 PAV sequences that were longer than 5 kb in B73 and Mo17, respectively. These PAV sequences were unevenly distributed across the genome (Fig. 1), and some were located in clusters (Supplementary Table 9). The longest PAV sequence segment was a 2.9-Mb B73-specific segment containing 66 predicted genes, found from 22.5 Mb to 25.4 Mb on chromosome 6. The length of this PAV segment was slightly longer than previously reported3. The longest Mo17-specific segment was a 2.5-Mb segment on chromosome 6 from 64.0 Mb to 66.5 Mb. This Mo17-specific segment was close to the centromere and contained only ten predicted genes. In addition, we found two Mo17-specific segments located close together on chromosome 2, with lengths of 752.6 kb (chromosome 2: 235809501–236562100) and 635.2 kb (chromosome 2: 237529501–238164700), containing 20 and 23 predicted genes, respectively (Supplementary Table 9). With the criterion requiring at least 75% of coding sequences to overlap with PAV sequences, and validation through alignment of resequencing reads, we identified 72 B73-specific PAV genes and 50 Mo17-specific PAV genes. The number of PAV genes identified with this stringent criterion was smaller than the earlier estimation using only resequencing data or comparative genomic hybridization arrays2,3,5. We also identified 22 PH207-specific PAV genes, as compared with B73, and 75 PH207-specific PAV genes, as compared with Mo17, by using the same method (Supplementary Table 10); only four genes overlapped between the two sets. We found that only ~25% of these PAV genes in the B73, Mo17 and PH207 genomes have likely orthologs in sorghum (Supplementary Table 10). To further trace the origin of these PAV genes, we aligned resequencing reads of 19 wild relatives, 23 landraces and 60 modern inbred lines from maize Hapmap2 (ref. 36) projects to the B73, Mo17 and PH207 genomes. Closely related homologs of more than 95% of these PAV genes were detected in at least one of the wild relatives, thus indicating that most of these PAV genes might have already existed in their direct ancestors (Supplementary Table 10). In summary, most of the PAV genes are present in the wild relatives of maize. These PAV genes might have arisen during the process of rediploidization of maize ancestors, which occurred before the completion of maize domestication but after the divergence of sorghum and maize. Thus, these PAV genes are now dispersed in landraces and modern maize lines (Supplementary Fig. 6).
A total of 9,867,466 SNPs and 1,422,446 indels were identified in the aligned syntenic blocks between the B73 and Mo17 genomes, with an average of 7.66 SNPs and 1.11 indels per kilobase (Supplementary Table 11). The distribution of SNPs and indels was positively correlated (Pearson’s correlation R = 0.76, P < 8.7 × 10−17; Fig. 1). Compared with indels genome wide, indels with multiples of 3 bp in length were more abundant in coding regions (Supplementary Fig. 7). We also identified 8,598,810 SNPs and 1,190,826 indels in the aligned syntenic sequences between the Mo17 and PH207 genomes, and 8,290,900 SNPs and 1,078,722 indels in the aligned syntenic sequences between the B73 and PH207 genomes (Supplementary Table 8).
Maize is an ancient tetraploid, and its two subgenomes have undergone extensive gene fractionation. Using fractionation bias estimation with sorghum as a reference, we found that the subgenome organizations of B73 and Mo17 were nearly identical (Supplementary Fig. 8), thus indicating that B73 and Mo17 share the same tetraploidization and a large part of the subsequent fractionation events. Fractionation of genes was similarly biased toward subgenome 1 in both B73 (23,029 and 14,877 genes for subgenomes 1 and 2, respectively) and Mo17 (22,471 and 14,366 genes for subgenomes 1 and 2, respectively), and subgenome 1 (B73, 1,206 Mb; Mo17, 1,205 Mb) had an approximately ~1.62-fold-longer physical length than that of subgenome 2 (B73, 740 Mb; Mo17, 745 Mb; Fig. 1 and Supplementary Fig. 8).
To detect genes that might be under selection, we first calculated the neutral mutation rate (Ks) between orthologous genes for each pair between any two genomes of B73, Mo17 and PH207. We found two peaks in the Ks distribution: one corresponded to a group of genes that might derive from recent genetic exchanges (Ks < 0.0028), and another represented most of the remaining genes that may have diverged from the common ancestors of maize approximately 2.1 Ma (Ks ~0.025) (Fig. 2a). We then calculated the Ka/Ks ratio for genes with Ks between 0.0028 and 0.25 (Fig. 2b). As expected, the Ka/Ks ratios for these genes were highly skewed toward zero, because most nonsynonymous mutations were deleterious and experienced strong purifying selection. Approximately 7,000 genes in each of the three genomes were identified to be likely to be evolutionary constrained (Ka/Ks < 0.1). In contrast, relatively few genes (>1,000) were detected to be under positive selection (Ka/Ks > 1).
Extensive gene-order and structural variations
Comparative analysis revealed 33,681 B73 and 33,597 Mo17 genes with corresponding orthologous genes or gene fragments in their syntenic regions. However, we found 5,105 B73 genes and 4,008 Mo17 genes that were nonsyntenic, because no homologous genes or gene fragments were found within 10 Mb of their corresponding positions in their respective counterpart genome (Table 2); these nonsyntenic genes accounted for 13.16% and 10.66% of the total analyzed genes in the B73 and Mo17 genomes, respectively. Similarly, 3,284 (9.02%) and 4,472 (12.27%) PH207 genes were defined as nonsyntenic genes when compared with B73 and Mo17 genomes, respectively (Supplementary Tables 12 and 13). Notably, 2,112 PH207 genes were nonsyntenic with both the B73 and Mo17 genomes.
Among the syntenic genes in B73 and Mo17 were 12,167 B73 and 12,674 Mo17 genes without any amino acid changes, and approximately 80% and 57% of genes showed no variation in coding sequences (CDSs) and gene bodies (CDS and intron), respectively (Table 2). We identified 2,498 B73 and 2,458 Mo17 highly conserved genes with no genetic variation in their entire genic regions, including within 2 Kb upstream and downstream (Table 2). There were 15,955 B73 and 15,512 Mo17 genes with only missense mutations and/or nonframeshift indels; those genes, together with the genes without amino acid changes, were classified as structurally conserved genes. These structurally conserved genes accounted for approximately 84% of the syntenic genes on the basis of either the B73 or the Mo17 gene annotation (Table 2). However, approximately 12% of the syntenic genes (3,947 in B73; 4,020 in Mo17) had large-effect mutations such as start- or stop-codon mutations, splice-donor or splice-acceptor mutations, frameshift mutations or premature-stop-codon mutations (Table 2). In addition, 1,612 B73 and 1,391 Mo17 genes had large structural variations, as compared with their corresponding syntenic genes in the Mo17 and B73 genomes, respectively (Table 2).
We then conducted additional analysis of the nonsyntenic genes between B73 and Mo17. On the basis of the analysis of a simple best hit in the counterpart genome, we classified 1,534 B73 and 1,216 Mo17 nonsyntenic genes as structurally conserved (Table 2). 1,387 B73 and 977 Mo17 nonsyntenic genes had large-effect mutations. In addition, 2,112 B73 and 1,765 Mo17 nonsyntenic genes had large structural variations, and 87 B73 genes and 253 Mo17 genes had no homologs identified in the Mo17 or B73 genome. Clustering of all annotated B73 and Mo17 genes revealed 320 B73-specific gene families (830 total gene members) as compared with Mo17, and 170 Mo17-specific gene families (578 gene members) as compared with B73. Among the 5,105 nonsyntenic genes in B73, 4,285 genes belonged to 2,114 gene families, including 294 B73-specific gene families (465 gene members). Among the 4,008 nonsyntenic genes in the Mo17 genome, 3,225 genes belonged to 1,631 gene families, including 119 Mo17-specific gene families (208 gene members).
In summary, a total of 9,058 B73 genes and 8,153 Mo17 genes had either large-effect mutations or large structural variations, as compared with their syntenic or best-hit Mo17 and B73 counterparts (Supplementary Table 13). Similarly, 8,278 and 9,738 PH207 genes had either large-effect mutations or large structural variations, as compared with the corresponding genes in B73 and Mo17, respectively (Supplementary Table 13). More than 20% of all predicted genes showed considerable protein sequence variations between any two inbred lines among B73, Mo17 and PH207, thus suggesting a potential functional complementation among these three representative maize inbred lines.
Notably, the proportion of genes with large-effect mutations and large structural variations within nonsyntenic genes was significantly higher than that in syntenic genes (chi-square test, P < 2 × 10−16). For example, the proportion of genes with large structural variations in the nonsyntenic gene group was with approximately ten times greater than that of the syntenic genes (Table 2 and Supplementary Table 12). Interestingly, some of the nonsyntenic genes in the B73 genome had very high sequence homology with genes in Mo17 that were classified as syntenic genes. In fact, only 971 B73 nonsyntenic genes that had their best hits in the Mo17 genome were also identified as nonsyntenic genes. Among them, 479 genes were identified as mutual nonsyntenic best hits between B73 and Mo17. Our results showed that 1,529 B73 nonsyntenic genes with their best-hit genes in Mo17 had syntenic homologs in B73. Similarly, 1,301 Mo17 nonsyntenic genes with their best-hit genes in B73 had syntenic homologs in Mo17, thus indicating that a large proportion of these nonsyntenic genes are members of multiple gene families.
To examine the extent of gene amplification, we clustered all annotated genes in the B73, Mo17 and PH207 genomes. As a result, 3,589 (9.1%) B73, 5,044 (13.1%) Mo17 and 4,987 (13.3%) PH207 genes were identified as singleton genes (Supplementary Table 14). Most of the genes were amplified through either dispersed duplication (20,874 B73 genes; 19,468 Mo17 genes; and 20,082 PH207 genes) or whole-genome duplication and segmental duplication (11,249 B73; 10,675 Mo17; and 9,908 PH207 genes; Supplementary Table 14). The remainder were grouped as proximal duplications or tandem duplications. We found one extreme case, a germin-like plant-defense-related gene37, that had 20 copies in B73 but only ten copies in Mo17 and nine copies in PH207 (Supplementary Fig. 9). The percentage of transcription factors in whole-genome-duplicated and segmental-duplicated genes was approximately tenfold higher than that in singleton genes, thus indicating that transcription factors tend to be retained after whole-genome duplication or segmental duplication (Supplementary Table 14), similarly to previous findings in Arabidopsis38. On the basis of the gene-clustering analysis, approximately 75% of nonsyntenic genes among B73, Mo17 and PH207 were members of gene families amplified via dispersed duplication (Supplementary Table 15). Further analysis showed that only approximately 11–23% of nonsyntenic genes among B73, Mo17 and PH207 had high-confidence syntenic orthologous genes in sorghum. In contrast, more than half (approximately 58%) of all annotated genes had syntenic orthologs in sorghum (Supplementary Table 16), thus suggesting that most of the nonsyntenic genes among B73, Mo17 and PH207 arose largely from gene amplification instead of differential fractionation between two subgenomes after tetraploidization.
To investigate the relationship between genomic variation and transcriptomic differences, we generated RNA-seq data for bract, root, stem, seedling and endosperm tissues from both B73 and Mo17. In total, 24,209 B73 genes and 23,947 Mo17 genes were expressed in at least one of these tissues (Supplementary Table 17), and 859 B73 genes and 770 Mo17 genes showed specific expression in at least one of the tissues, including 25 B73-specific and 16 Mo17-specific PAV genes (Supplementary Table 18). We also found significant differences in expression profiles among different categories of genes that varied in their level of conservation. 66% of structurally conserved genes were expressed, a higher value than the proportion of genes with large-effect mutations and large structural variation (average 48%). Moreover, approximately 1.4% of structurally conserved genes were specifically expressed in B73 or Mo17, a value significantly lower than the proportion of genes with large-effect mutations and large structural variations (average 4.5%) (Supplementary Table 17).
Discussion
The availability of high-quality assembled genomes for both B73 and Mo17 provides an unprecedented opportunity for extensive intraspecific genome comparison in maize. Direct comparison between the two maize genomes uncovered extensive SNP and indel variation, as well as a large extent of gene-order and structural variations. We demonstrated that more than 10% of B73 and Mo17 genes were mutually nonsyntenic, a value approximately two- to threefold higher than the proportion of nonsyntenic genes between R498 (indica) and Nipponbare (japonica) Asian rice (Supplementary Tables 13 and 19). Although some of the nonsyntenic genes have closely related homologs between these two maize lines, the positional changes of these genes may introduce different chromatin contexts affecting the expression of the genes themselves or their neighboring genes. The exact functions of these nonsyntenic genes are largely unknown, because they are underrepresented in classical genetics studies in maize39. These nonsyntenic genes may play important roles in some lineage-specific functions, as demonstrated in a study on root development40. Evaluating the contribution of these nonsyntenic genes to quantitative phenotypic variations of agronomic traits would be an interesting future pursuit.
In addition to the gene-order variations, there were several other types of intraspecific structural variations. Only 60% of the B73 and Mo17 genomes were able to be aligned as one-to-one blocks. Although the remaining 40% of the variable genome largely comprised repetitive elements, the B73 and Mo17 genomes each contained approximately 12 Mb of unique low-copy sequences, including 122 (72 in B73 and 50 in Mo17) high-stringency PAV genes. Furthermore, more than 20% of the annotated genes had large-effect mutations or large structural variations, which could potentially lead to protein sequence changes and potential functional divergence between the two maize lines. Even after exclusion of potential redundancy resulting from multigene families, there were 320 B73-specific (and 170 Mo17-specific) gene families. In addition, there were 859 B73 genes and 770 Mo17 genes that showed specific expression in at least one of the tissues tested.
Several factors can contribute to the extensive intraspecific genome diversity observed. Transposable elements, such as helitrons, have been reported to be able to introduce gene movement or exon shuffling in the maize genome41,42. The extensive genome diversity found suggests that the hybrids generated between two different maize lines may have a dramatically different complement of proteins or regulatory sequences, thus supporting the complementation hypothesis explaining the exceptional heterosis observed in maize nearly a century ago7,43,44.
URLs
Falcon, https://github.com/PacificBiosciences/FALCON/; Arrow, https://github.com/PacificBiosciences/GenomicConsensus/; blasr, https://github.com/PacificBiosciences/blasr/; Bwa, http://bio-bwa.sourceforge.net/; Pilon, https://github.com/broadinstitute/pilon/; IrysSolve, https://bionanogenomics.com/support/software-downloads/; GenBank, https://www.ncbi.nlm.nih.gov/genbank/; RepeatModeler, http://www.repeatmasker.org/RepeatModeler/; RepeatMasker, http://www.repeatmasker.org/RMDownload.html; Repbase, http://www.girinst.org/repbase/; LTRharvest, http://genometools.org/index.html; LTRdigest, http://genometools.org/index.html; GyDB, http://gydb.org/; MUSCLE, https://www.drive5.com/muscle/; EMBOSS, http://emboss.sourceforge.net/; distmat, http://emboss.sourceforge.net/apps/release/6.6/emboss/apps/distmat.html; MAKER-P, http://www.yandell-lab.org/software/maker-p.html; Augustus, http://bioinf.uni-greifswald.de/augustus/; FGENESH, http://www.softberry.com/berry.phtml/; Mummer, http://mummer.sourceforge.net/; Blastp, https://blast.ncbi.nlm.nih.gov/Blast.cgi/; MCscanX, https://github.com/wyp1125/MCScanX/; Phytozome, http://www.phytozome.net/; OrthoMCL, http://orthomcl.org/orthomcl/; Synmap, https://genomevolution.org/coge/; last, http://last.cbrc.jp/; DAGchainer, http://dagchainer.sourceforge.net/; Quota Align, https://github.com/tanghaibao/quota-alignment/; PAML, http://abacus.gene.ucl.ac.uk/software/paml.html.
Methods
Plant material
The maize (Z. mays) inbred line Mo17 was selected for sequencing because of its important role in maize breeding and genetic research. The plants were grown in a greenhouse at 25 °C in dark conditions for 14 d, and the aerial parts of seedlings were harvested and frozen immediately in liquid nitrogen for extraction of genomic DNA. High-molecular-weight genomic DNA for library construction was extracted from isolated nuclei.
PacBio and Illumina library construction and sequencing
Libraries for SMRT PacBio genome sequencing were constructed as described previously45. Approximately 20 µg of high-quality genomic DNA was sheared to an ~20 -kb targeted size and assessed with an Agilent 2100 Bioanalyzer. Shearing of genomic DNA was followed by damage repair and end repair, blunt-end adaptor ligation and size selection with a Blue Pippin system (Sage Science). The final libraries were sequenced on the PacBio Sequel platform (Pacific Biosciences).
Libraries for Illumina PCR-free paired-end genome sequencing were constructed according to the standard manufacturer’s protocol (Illumina). Approximately 5 µg genomic DNA was fragmented, then size-selected (450 bp and 800 bp) through agarose gel electrophoresis. The ends of selected DNA fragments were blunted with an A-base overhang and ligated to sequencing adapters. After quality control, all the PCR-free libraries were sequenced on an Illumina platform with a paired-end sequencing strategy.
De novo assembly of PacBio SMRT reads
Approximately 24 million PacBio SMRT reads were used for contig assembly with Falcon45. All reads were first pairwise compared, and sequencing errors were corrected with parameters ‘--length_cutoff 12000 --length_cutoff_pr 14000’. Preassembly and further error correction were then performed with parameters ‘--min_cov 4 --max_n_read 300’. The error rate used for overlapping detection was ‘--e 0.96’, and overlaps were filtered with the parameters ‘--bestn 10 --max_cov 60 --max_cov 2 --max_diff 60’. Overlapping graphs were constructed, and contigs were finally generated according to these graphs. All PacBio SMRT reads were then mapped back to the contigs with Blasr46 with the parameters ‘--bestn 10 --minMatch 12 --minSubreadLength 500 --minAlnLength 500 --minPctSimilarity 70 --minPctAccuracy 70 --hitPolicy randombest --randomSeed 1’. Arrow (see URLs) was used to correct the sequencing errors with default parameters according to the alignments. The Illumina PCR-free paired-end reads were mapped to the corrected contigs above with BWA mem47 with default parameters, and high-quality mapped reads (MAQ >20) were further used to polish the assembly with Pilon24 with default parameters. This procedure resulted a total assembly length of 2.15 Gb with an N50 length of 1.48 Mb.
Construction of BioNano optical maps
High-molecular-weight DNA was isolated from the same tissue as described in the ‘Plant material’ section, digested by the single-stranded nicking endonuclease Nt.BspQI and then labeled with IrysPrep Labeling mix and Taq polymerase according to standard BioNano protocols. Labeled DNA was imaged automatically with the BioNano Irys system. BioNano raw BNX files were de novo assembled into genome maps with IrysSolve (see URLs). All single molecules were first sorted and autodenoised. Pairwise comparison was performed with RefAligner (see URLs) to identify all molecule overlaps, and consensus maps were constructed. All molecules were then mapped back to the consensus maps, and the maps were recursively refined and extended (five times). The final genome maps were 2.24 Gb in total, and the N50 of the maps was 1.41 Mb.
Hybrid assembly of PacBio contigs and BioNano optical maps
The BioNano IrysSolve (see URLs) module ‘HybridScaffold’ was used to perform the hybrid assembly between PacBio-assembled contigs and BioNano-assembled genome maps. The main procedure was as follows: PacBio contigs were first converted into cmap format. BioNano cmaps were then aligned to the contig cmaps with RefAligner, and this was followed by label rescaling. The rescaled BioNano cmaps were aligned again to the contig cmaps, and sequences were split at the conflict points. The ‘aggressive’ configuration was used, and scaffolds were built according to the alignment information between PacBio contigs and BioNano genome maps. There were 2,116 PacBio contigs with a total length of 2.08 Gb linked by BioNano maps, which resulted in 348 scaffolds with an N50 of 10.51 Mb, with a total length of 2.12 Gb and ~40-Mb gaps introduced. The total length of the hybrid scaffolds and unscaffolded PacBio contigs was 2.18 Gb with an N50 of 10.42 Mb.
Assembly evaluation
We used published Mo17 BAC sequences to assess the quality of the hybrid assembly. Nine Mo17 BAC sequences (1,528 kb) downloaded from GenBank (see URLs) were mapped to the scaffolds with BLAT26, and the alignment results were manually checked. All nine Mo17 BACs were covered by a single scaffold, with 100% coverage and 99.97% identity of the total BAC sequences, thus suggesting that the genome assembly was of high quality.
BWA mem47 was used to map the ~4.4 million maize GBS-tag sequences27 to the genome sequences to evaluate the assembly. The alignments were further filtered by mapping quality (MAQ = 60), and 36.68% were kept. Scaffolds with more than ten tags aligned were evaluated, and three scaffolds with tags aligned at disparate chromosome locations were split at the appropriate gap positions. After this correction, there were 2,560 scaffolds with an N50 length of 10.2 Mb, and the total genome length was 2.18 Gb. BUSCO25 was further used to evaluate the genome-assembly completeness. ‘Embryophyta_odb9’, which contained 1,440 single-copy orthologous genes was used as a searching dataset, and both Mo17 and B73 genomes were assessed.
Construction of pseudomolecules
The maize pangenome GBS tags27 were also used to anchor the assembled scaffolds onto Mo17 chromosomes. According to the mapping results above, scaffolds with more than ten tags aligned were further used to construct the pseudomolecules. The order and orientation of scaffolds were determined according to the physical positions of GBS tags. In total, 2.1-Gb scaffold sequences accounting for 96.4% of the Mo17 assembled genome were anchored onto the ten Mo17 chromosomes, which contained 97.95% of the annotated genes. We also aligned all GBS-tag sequences to the B73 genome and compared the alignments in these two genomes. The alignments showed high consistency between two genomes except for some pericentromere regions, owing to the lack of GBS tags.
Analysis of repetitive elements
We identified repetitive elements in the Mo17 genome through a combination of homolog-based and de novo approaches. RepeatModeler48 was first used to build TE consensus sequences as a de novo TE library on the basis of the Mo17 genome sequence. RepeatMasker49 was then used to discover and identify repeats in the Mo17 genome with the combined library of the de novo TEs of Mo17 and Repbase50. Repetitive elements in the B73 and PH207 genomes were identified through the same method.
Transposon activity analysis
Full-length LTR retrotransposons were first identified in the assembled sequences of the B73, Mo17 and PH207 genomes with LTRharvest51 with the following parameters: ‘-longoutput -motif tgca -minlenltr 100 -maxlenltr 7000 -mindistltr 1000 -maxdistltr 20000 -similar 85 -motifmis 1 -mintsd 5 -xdrop 5 -overlaps best’. All predicted LTR retrotransposons were further annotated for protein domains with LTRdigest52 with GyDB (see URLs) as a search database. Candidates with no typical protein domains (for example, GAG, INT, RT and RT) or a tandem-repeat content greater than 20% were filtered. This procedure resulted in a final set of 73,459, 74,160 and 50,402 high-confidence full-length LTR retrotransposons in the B73, Mo17 and PH207 genomes, respectively. To calculate the insertion age of each LTR retrotransposons, 5′ and 3′ LTRs of the same element were aligned with MUSCLE53, and the distmat utility in the EMBOSS54 software package was used to calculate the accumulated divergence ‘K’ between 5′ and 3′ LTRs. Insertion times (T) of the LTR retrotransposons were calculated with the formula T = K/2 × r, where r is the TE-specific mutation rate of 1.3 × 10−8 per site per year55.
RNA-seq data collection and generation
To aid in genome annotation and to perform transcriptome analysis, we generated RNA-seq data for five different tissues from B73 and Mo17: endosperm 12 d after pollination, 14-d seedlings, bracts, roots and stems harvested in the silking stage. For each sample, two independent biological replicates were generated. All fresh tissues were frozen in liquid nitrogen and stored at –80 °C before processing. Total RNA of each sample was extracted with TRIzol according to the manufacturer’s instructions. RNA-seq libraries were prepared with the Illumina standard mRNA-seq library preparation kit and sequenced on the Illumina platform with paired-end sequencing strategy.
Gene annotation
MAKER-P version 3.1 (ref. 56) was used to annotate genes in the Mo17 genome, through a comprehensive strategy combining results obtained from protein-homology-based prediction, RNA-seq-based prediction and ab initio prediction. We used the same evidence that was used for the previous B73 gene annotation, with the addition of Mo17-specific RNA-seq datasets. All annotated proteins from Sorghum bicolor, Oryza sativa, Setaria italica, Brachypodium distachyon and Arabidopsis thaliana, downloaded from http://gramene.org/ release 48 (ref. 57), were used for protein-homology-based prediction. 74,471 assembled transcripts from multiple Mo17 tissues, full-length transcripts from B73 Iso-seq58, another set of 69,163 publicly available full-length cDNAs from B73 deposited in GenBank59, a total of 1,574,442 Trinity-assembled transcripts from 94 B73 RNA-seq experiments33 and 112,963 transcripts assembled from deep sequencing of a B73 seedling60 were collected and included as transcript evidence. Augustus61 and FGENESH (see URLs) were used for ab initio prediction of gene models in the TE-masked Mo17 genome. 44,747 genes (53,021 transcripts) were identified in the Mo17 genome and are referred to as the working gene set. This working set of gene annotations was expected to contain TEs that were not masked before annotation or annotations with poor supporting evidence. We further filtered this working set according to AED scores generated in MAKER-P software, then confirmed splice sites and performed transposon screening. Finally, 38,620 high confidence genes remained and are referred to as the filter gene set.
Identification of SNPs and indels
We identified SNPs and insertion/deletion polymorphisms (indels, length <100 bp) between the B73 and Mo17 genomes with Mummer34 as follows: (i) The Mo17 pseudochromosome sequence was mapped to its corresponding B73 pseudochromosome with nucmer with the parameters ‘-mumreference -g 1000 -c 90 -l 40’. (ii) The delta-filter was used to filter mapping noise and determine the one-to-one alignment blocks with parameters ‘-r -q’. Alignments with aligned positions in one genome that were located more than 10 Mb away in another genome were further filtered. The aligned blocks between these two genomes were identified, and blank regions on the chromosomes that might be low-similarity regions or multiple aligned regions were filtered. (iii) Show-snps was used to obtain SNPs and small indels (<100 bp). B73-genome-based SNPs and indels were detected with the parameter ‘-ClrTH’, and Mo17-genome-based parameters were detected with the parameter ‘-ClqTH’. SNPs and indels shared between the Mo17 and PH207 genomes, or the B73 and PH207 genomes, were processed with the same method used for SNPs shared between the B73 and Mo17 genomes. The genome distributions of SNPs and indels between the B73 and Mo17 genomes were also determined.
Identification of PAV sequences, PAV clusters and PAV genes
PAV sequences in the B73 and Mo17 genomes were identified through a sliding-window method. To identify B73 specific sequences, we divided the B73 genome into 500-bp overlapping windows with a step size of 100 bp and then aligned each 500-bp window against the B73 and Mo17 genomes with BWA mem47 with options ‘-w 500 --M’. The sequences of windows that could not be aligned or that aligned to the Mo17 genome with a primary alignment coverage less than 25% but could be properly aligned to the B73 genome were defined as B73-specific sequences. Overlapping windows that could not be aligned were merged. Mo17- and PH207-specific sequences were identified through the same method. Most of these PAV sequences had a relatively short length (Supplementary Fig. 5), possibly as a result of our stringent calling criteria. We further merged PAV sequences that were within 100 kb of the physical coordinates to identify PAV clusters. If a merged region had more than 10% PAV sequences, we defined this region as a PAV cluster. We listed some large PAV clusters (>500 kb) in both the B73 and Mo17 genomes.
For the identification of B73-, Mo17- and PH207-specific genes, the CDS of different transcripts were merged to represent a single gene, and genes with more than 75% of the CDS regions covered by PAV sequences were defined as PAV genes. We further aligned B73 resequencing reads from maize HapMap2 (ref. 36) projects to the Mo17 genome and Mo17 Illumina PCR-free reads to the B73 genome with BWA mem47 to exclude potential false positives. For the B73/Mo17-specific genes above, we filtered those with more than 50% CDS regions covered by Mo17/B73 reads to obtain the final PAV genes.
Resequencing reads of 19 wild relatives, 23 landraces and 60 modern inbred lines from maize Hapmap2 (ref. 36) projects were aligned to the B73, Mo17 and PH207 genomes with Bwa mem47. The mapping depth across the PAV gene regions was calculated with samtools62. If a gene had more than 90% of the coding sequences covered by resequencing reads, we defined it as trackable in that line.
Detection of gene structural variations among B73, Mo17 and PH207
To survey gene-structure variation between B73 and Mo17 genomes, we extended the longest transcript of each B73/Mo17 gene 2 kb upstream and downstream, and then aligned it to both Mo17/B73 genomes and the sequence from the Mo17/B73 syntenic region (in which the locations in the two genomes were less than 10 Mb apart), respectively, with Bwa mem47. The alignments separated by less than 20 kb were merged. For the best-hit-based method, the genome-wide best hits were used to assess gene-structure variation. Genes without amino acid substitutions or with only missense mutations and/or nonframeshift indels (length = 3n bp) were classed as structurally conserved genes. Genes with complete CDSs but containing SNPs or indels (3 ± 1 nt) that might produce initiation codons, termination codons, premature termination, splicing-donor-site or splicing-acceptor-site mutations, and ORF frameshifts were classified as genes with large-effect mutations. The remaining genes not identified as PAV genes were classified as genes with large structural variation.
For the synteny-based method, the best hits located in the syntenic regions were used to assess gene-structure variation. Structurally conserved genes and genes with large-effect mutations were defined according to the same criteria above. The remaining genes with more than 75% CDS missed in the syntenic regions were classed as genes with syntenic information not established or were otherwise classified as genes with large structural variation. Notably, only genes and their best hits in the counterpart genome anchored in ten pseudomolecules were included in the analysis. The comparisons of PH207 genes to the B73 and Mo17 genomes were performed in the same way.
Identification of duplicated genes and gene families
To identify gene duplications, BLASTP63 was used to calculate pairwise similarities (e value < 1 × 10−20), and MCscanX64 with default parameters was then used for classification.
To identify gene families, we merged annotated genes from Mo17, B73 and three other grasses from the Phytozome database (see URLs), including S. bicolor (33,032 genes), O. sativa (39,049 genes), B. distachyon (31,694 genes) and A. thaliana (27,416 genes). The longest proteins for each gene were aligned to one another. BLASTP63 was used to calculate pairwise similarities (e value < 1 × 10−20), and OrthoMCL65 was used to identify gene families with an inflation value of 2 and percent-match cutoff of 50.
Comparative genomic analysis among B73, Mo17, PH207 and sorghum
To perform the comparative genomic analysis, we used the Synmap pipeline (see URLs). In brief, we used last66 to blast the CDS sequence, then detected syntenic blocks with DAGchainer67 with options -D 20 -A 5. Quota Align68 was further used to merge adjacent syntenic blocks. The syntenic depth was set to 2:1 for maize and sorghum and 1:1 for B73 and Mo17; B73 and PH207; and Mo17 and PH207 comparisons, and the overlapped distance was set to 40 to permit overlapped syntenic regions. Fractionation bias was applied to determine subgenome organization in maize compared with sorghum. The CodeML utility in the PAML69 software package was used to calculate the Ka and Ks rates between orthologous genes. The time of divergence from the common ancestors of maize (~2.1 Ma) was inferred on the basis of the Ks of 0.025, because maize and sorghum shared a common ancestor with ~11.9 Ma (Ks ~0.139).
Transcriptome comparison between B73 and Mo17
The RNA-seq data for the bract, root, stem, seedling and endosperm tissues from the B73 and Mo17 lines were used to perform transcriptome comparison between B73 and Mo17. The RNA-seq data were aligned to both the B73 and Mo17 genomes with Hisat2 (ref. 70). All aligned reads were used to calculate the fragments per kilobase per million (FPKM) values with Cufflinks71. Genes with FPKM value greater than 1 for tissues of B73 (Mo17) and lower than 0.1 in corresponding tissues of Mo17 (B73) were deemed B73 (Mo17) specifically expressed genes.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Code availability
Custom codes used in this study are currently hosted in a GitHub repository at https://github.com/caulai/Mo17_genome_assembly/.
Data availability
The genome assembly and gene annotation have been deposited in the NCBI database under BioProject number PRJNA358298 and BioSample number SAMN06169745. The GenBank accession number of the above data is NCVQ00000000. Raw PacBio SMRT reads, Illumina data and RNA-seq data have been deposited in the NCBI SRA database under accession number SRP111315.
References
Beló, A. et al. Allelic genome structural variations in maize detected by array comparative genome hybridization. Theor. Appl. Genet. 120, 355–367 (2010).
Lai, J. et al. Genome-wide patterns of genetic variation among elite maize inbred lines. Nat. Genet. 42, 1027–1030 (2010).
Springer, N. M. et al. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5, e1000734 (2009).
Buckler, E. S., Gaut, B. S. & McMullen, M. D. Molecular and functional diversity of maize. Curr. Opin. Plant Biol. 9, 172–176 (2006).
Swanson-Wagner, R. A. et al. Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 20, 1689–1699 (2010).
Schnable, P. S. et al. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115 (2009).
Duvick, D. N. Biotechnology in the 1930s: the development of hybrid maize. Nat. Rev. Genet. 2, 69–74 (2001).
Troyer, A. F. Background of US hybrid corn. Crop Sci. 39, 601–626 (1999).
Bortiri, E., Jackson, D. & Hake, S. Advances in maize genomics: the emergence of positional cloning. Curr. Opin. Plant Biol. 9, 164–171 (2006).
Stupar, R. M. & Springer, N. M. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173, 2199–2210 (2006).
Zhang, M. et al. Genome-wide high resolution parental-specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize. Genome Res. 24, 167–176 (2014).
Zhang, M. et al. Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc. Natl Acad. Sci. USA 108, 20042–20047 (2011).
Waters, A. J. et al. Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23, 4221–4233 (2011).
Eichten, S. R. et al. B73-Mo17 near-isogenic lines demonstrate dispersed structural variation in maize. Plant Physiol. 156, 1679–1690 (2011).
Edwards, D., Batley, J. & Snowdon, R. J. Accessing complex crop genomes with next-generation sequencing. Theor. Appl. Genet. 126, 1–11 (2013).
Hirsch, C. N. et al. Draft assembly of elite inbred line PH207 provides insights into genomic and transcriptome diversity in maize. Plant Cell 28, 2700–2714 (2016).
Dong, J. et al. Analysis of tandem gene copies in maize chromosomal regions reconstructed from long sequence reads. Proc. Natl Acad. Sci. USA 113, 7949–7956 (2016).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Du, H. et al. Sequencing and de novo assembly of a near complete indica rice genome. Nat. Commun. 8, 15324 (2017).
Mock, T. et al. Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 541, 536–540 (2017).
Bickhart, D. M. et al. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet. 49, 643–650 (2017).
Jarvis, D. E. et al. The genome of Chenopodium quinoa. Nature 542, 307–312 (2017).
Jiao, Y. et al. Improved maize reference genome with single-molecule technologies. Nature 546, 524–527 (2017).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963 (2014).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Kent, W. J. BLAT: the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Lu, F. et al. High-resolution genetic mapping of maize pan-genome sequence anchors. Nat. Commun. 6, 6914 (2015).
Feschotte, C., Jiang, N. & Wessler, S. R. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3, 329–341 (2002).
SanMiguel, P., Gaut, B. S., Tikhonov, A., Nakajima, Y. & Bennetzen, J. L. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20, 43–45 (1998).
Clark, R. M., Tavaré, S. & Doebley, J. Estimating a nucleotide substitution rate for maize from polymorphism at a major domestication locus. Mol. Biol. Evol. 22, 2304–2312 (2005).
Baucom, R. S. et al. Exceptional diversity, nonrandom distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 5, e1000732 (2009).
Luo, M. C. et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551, 498–502 (2017).
Law, M. et al. Automated update, revision, and quality control of the maize genome annotations using MAKER-P improves the B73 RefGen_v3 gene models and identifies new genes. Plant Physiol. 167, 25–39 (2015).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004).
Brunner, S., Fengler, K., Morgante, M., Tingey, S. & Rafalski, A. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17, 343–360 (2005).
Chia, J. M. et al. Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44, 803–807 (2012).
Breen, J. & Bellgard, M. Germin-like proteins (GLPs) in cereal genomes: gene clustering and dynamic roles in plant defence. Funct. Integr. Genom. 10, 463–476 (2010).
Blanc, G. & Wolfe, K. H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16, 1679–1691 (2004).
Salvi, S. An evo-devo perspective on root genetic variation in cereals. J. Exp. Bot. 68, 351–354 (2017).
Tai, H. et al. Nonsyntenic genes drive RTCS-dependent regulation of the embryo transcriptome during formation of seminal root primordia in maize (Zea mays L.). J. Exp. Bot. 68, 403–414 (2017).
Morgante, M. et al. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 37, 997–1002 (2005).
Lai, J., Li, Y., Messing, J. & Dooner, H. K. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc. Natl Acad. Sci. USA 102, 9068–9073 (2005).
Fu, H. & Dooner, H. K. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl Acad. Sci. USA 99, 9573–9578 (2002).
Paschold, A. et al. Nonsyntenic genes drive highly dynamic complementation of gene expression in maize hybrids. Plant Cell 26, 3939–3948 (2014).
Pendleton, M. et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods 12, 780–786 (2015).
Chaisson, M. J. & Tesler, G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13, 238 (2012).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
Smit, A. & Hubley, R. RepeatModeler Open-1.0. http://www.repeatmasker.org/RepeatModeler/ (2010).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinforma. 25, 4.10 (2009).
Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005).
Ellinghaus, D., Kurtz, S. & Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9, 18 (2008).
Steinbiss, S., Willhoeft, U., Gremme, G. & Kurtz, S. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 37, 7002–7013 (2009).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Rice, P., Longden, I. & Bleasby, A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000).
Ma, J. & Bennetzen, J. L. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl Acad. Sci. USA 101, 12404–12410 (2004).
Cantarel, B. L. et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196 (2008).
Tello-Ruiz, M. K. et al. Gramene 2016: comparative plant genomics and pathway resources. Nucleic Acids Res. 44, D1133–D1140 (2016).
Wang, B. et al. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 7, 11708 (2016).
Soderlund, C. et al. Sequencing, mapping, and analysis of 27,455 maize full-length cDNAs. PLoS Genet. 5, e1000740 (2009).
Martin, J. A. et al. A near complete snapshot of the Zea mays seedling transcriptome revealed from ultra-deep sequencing. Sci. Rep. 4, 4519 (2014).
Keller, O., Kollmar, M., Stanke, M. & Waack, S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27, 757–763 (2011).
Li, H. et al. The sequence alignment/map format and samtools. Bioinformatics 25, 2078–2079 (2009).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Wang, Y. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012).
Li, L., Stoeckert, C. J. Jr & Roos, D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003).
Kiełbasa, S. M., Wan, R., Sato, K., Horton, P. & Frith, M. C. Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487–493 (2011).
Haas, B. J., Delcher, A. L., Wortman, J. R. & Salzberg, S. L. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics 20, 3643–3646 (2004).
Tang, H. et al. Screening synteny blocks in pairwise genome comparisons through integer programming. BMC Bioinformatics 12, 102 (2011).
Yang, Z. & Yang, Z. H. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Acknowledgements
We thank the Beijing Computing Center for providing computing resources. This work was supported by grants from the National Key Research and Development Program of China (2016YFD0100802) and the National Natural Science Foundation of China (91435206; 31421005) to J.L., and the US National Science Foundation (IOS-1238014 and USDA-ARS) to E.S.B.
Author information
Authors and Affiliations
Contributions
J.L. designed the research. S.S., Y.Z. and Hainan Zhao performed genome assembly and genome annotation. S.S., Y.Z., J.P.S. and Hainan Zhao performed genome comparison. Haiming Zhao, J.C., W.S. and Y.C. prepared DNA/RNA samples and constructed the next-generation-sequencing library. J.L., S.S., Y.Z., J.C., J.S., Haiming Zhao, Hainan Zhao, W.S., M.Z., X.D., H.L. and X.M. participated in the analysis. Y.J., B.W., X.W., J.C.S., D.H.W. and C.L. were involved in gene annotation. J.C.G., F.L. and E.S.B. provided the anchored GBS tags. G.Y. was involved in the PacBio data generation. K.F., B.L., A.R. and P.S.S. participated in discussion on genome assembling and validation. J.L., S.S., Y.Z., J.C. and J.P.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Comparison of the GBS tags aligned to the assembled sequences of B73 and Mo17 genomes.
The green blocks indicate pericentromere regions in B73 genome. The blue lines indicate aligned GBS tags to the two genomes
Supplementary Figure 2 The distribution of insertion ages of high-confidence full-length LTR retrotransposons in B73, Mo17 and PH207 genomes.
X axis represents insertion ages of LTR retrotransposons in the genome and Y axis represents the number of LTRs
Supplementary Figure 3 The chromosomal distribution of insertion ages of high-confidence full-length LTR retrotransposons in B73 and Mo17 genomes.
The insertion date of each LTR retrotransposon was averaged in 1Mb bin window. The black blocks indicate pericentromere regions in B73 genome
Supplementary Figure 4 Whole-genome comparison between B73 and Mo17.
Blue lines represented the one-to-one aligned regions between each pair of pseudomolecules
Supplementary Figure 5
Length distribution of PAV sequences between B73 and Mo17 genomes
Supplementary Figure 6 Coverage distribution of B73-, Mo17- and PH207-specific genes in 19 wild relatives, 23 landraces and 60 modern inbred lines.
For each gene, the coverage of CDS region was shown
Supplementary Figure 7
Histogram of indel numbers between B73 and Mo17 of the whole genome and coding regions
Supplementary Figure 8 Subgenome organization between B73, Mo17 and sorghum.
(a), B73 and Sorghum; (b), Mo17 and Sorghum. The red circle represents subgenome 1 and green circle represents subgenome 2
Supplementary Figure 9 Graphic representation of the alignment and polarity of homologous genes among B73, Mo17 and PH207 genomes.
Green blocks represent the homologs of germin-like plant defense related genes. Blue and red blocks indicate flank genes in each genome
Supplementary Information
Supplementary Text and Figures
Supplementary Figures 1–9
Supplementary Tables 1–19
Supplementary Tables 1–19
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, S., Zhou, Y., Chen, J. et al. Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat Genet 50, 1289–1295 (2018). https://doi.org/10.1038/s41588-018-0182-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0182-0
This article is cited by
-
A chromosome level genome assembly of Pseudoroegneria Libanotica reveals a key Kcs gene involves in the cuticular wax elongation for drought resistance
BMC Genomics (2024)
-
Cytoplasmic genome contributions to domestication and improvement of modern maize
BMC Biology (2024)
-
Plant pangenomes for crop improvement, biodiversity and evolution
Nature Reviews Genetics (2024)
-
Hybrid allele-specific ChIP-seq analysis identifies variation in brassinosteroid-responsive transcription factor binding linked to traits in maize
Genome Biology (2023)
-
Transcriptome-wide identification and characterization of genes exhibit allele-specific imprinting in maize embryo and endosperm
BMC Plant Biology (2023)