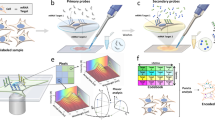
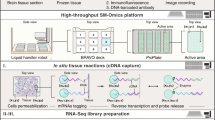
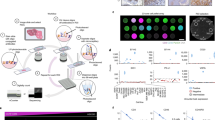
Abstract
Resolving the spatial distribution of RNA and protein in tissues at subcellular resolution is a challenge in the field of spatial biology. We describe spatial molecular imaging, a system that measures RNAs and proteins in intact biological samples at subcellular resolution by performing multiple cycles of nucleic acid hybridization of fluorescent molecular barcodes. We demonstrate that spatial molecular imaging has high sensitivity (one or two copies per cell) and very low error rate (0.0092 false calls per cell) and background (~0.04 counts per cell). The imaging system generates three-dimensional, super-resolution localization of analytes at ~2 million cells per sample. Cell segmentation is morphology based using antibodies, compatible with formalin-fixed, paraffin-embedded samples. We measured multiomic data (980 RNAs and 108 proteins) at subcellular resolution in formalin-fixed, paraffin-embedded tissues (nonsmall cell lung and breast cancer) and identified >18 distinct cell types, ten unique tumor microenvironments and 100 pairwise ligand–receptor interactions. Data on >800,000 single cells and ~260 million transcripts can be accessed at http://nanostring.com/CosMx-dataset.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The full RNA NSCLC dataset used in this study is available at http://nanostring.com/CosMx-dataset.
Code availability
Data from this publication (the full RNA NSCLC dataset) has been placed in the public domain in a format that can be analyzed and visualized using a variety of open-source packages, such as Seurat (https://github.com/satijalab/seurat) and Giotto (https://github.com/RubD/Giotto). Nearly all of the analyses performed in this paper can be accomplished using these open-source packages. For any of the specialized code/analyses performed in this manuscript that are not available through open-source packages, requests can be made through email to the corresponding author.
References
Garon, E. B. et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 372, 2018–28 (2015).
Yu, H. et al. PD-L1 expression by two complementary diagnostic assays and mRNA in situ hybridization in small cell lung cancer. J. Thorac. Oncol. 12, 110–120 (2017).
Yu, H., Boyle, T. A., Zhou, C., Rimm, D. L. & Hirsch, F. R. PD-L1 expression in lung cancer. J .Thorac. Oncol. 11, 964–975 (2016).
Ting, D. T. et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331, 593–596 (2011).
Garber, K. Oncologists await historic first: a pan-tumor predictive marker, for immunotherapy. Nat. Biotechnol. 35, 297–298 (2017).
Sokolenko, A. P. & Imyanitov, E. N. Molecular tests for the choice of cancer therapy. Curr. Pharm. Des. 23, 4794–4806 (2017).
Dereli, A. S., Bailey, E. J. & Kumar, N. N. Combining multiplex fluorescence in situ hybridization with fluorescent immunohistochemistry on fresh frozen or fixed mouse brain sections. J. Vis. Exp. https://doi.org/10.3791/6170910.3791/61709 (2021).
Taube, J. M. et al. The Society for Immunotherapy of Cancer statement on best practices for multiplex immunohistochemistry (IHC) and immunofluorescence (IF) staining and validation. J. Immunother. Cancer 8, e000155 (2020).
Hirsch, F. R. et al. PD-L1 immunohistochemistry assays for lung cancer: results from Phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. J. Thorac. Oncol. 12, 208–222 (2017).
Udall, M. et al. PD-L1 diagnostic tests: a systematic literature review of scoring algorithms and test-validation metrics. Diagn. Pathol. 13, 12 (2018).
Halse, H. et al. Multiplex immunohistochemistry accurately defines the immune context of metastatic melanoma. Sci. Rep. 8, 11158 (2018).
Macosko Evan, Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 161, 1202–1214 (2015).
Zilionis, R. et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc. 12, 44–73 (2017).
Wang, X., He, Y., Zhang, Q., Ren, X. & Zhang, Z. Direct comparative analyses of 10X Genomics Chromium and Smart-seq2. Genomics Proteomics Bioinformatics 19, 253–266 (2021).
See, P., Lum, J., Chen, J. & Ginhoux, F. A single-cell sequencing guide for immunologists. Front. Immunol. 9, 2425 (2018).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Stickels, R. R. et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 39, 313–319 (2021).
Fu, X. et al. Continuous polony gels for tissue mapping with high resolution and RNA capture efficiency. Preprint at bioRxiv https://doi.org/10.1101/2021.03.17.435795 (2021).
Liu, Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183, 1665–1681 (2020).
Merritt, C. R. et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 38, 586–599 (2020).
Wu, Q. et al. Poly A – transcripts expressed in HeLa cells. PLoS ONE 3, e2803 (2008).
Moffitt, J. R. et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl Acad. Sci. USA 113, 11046–11051 (2016).
Moffitt, J. R. & Zhuang, X. in Methods in Enzymology (eds Filonov, G. S. & Jaffrey, S. R.) 1–49 (Academic Press, 2016).
Groiss, S. et al. Highly resolved spatial transcriptomics for detection of rare events in cells. Preprint at bioRxiv https://doi.org/10.1101/2021.10.11.463936 (2021).
Lee, J. H. et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc. 10, 442–458 (2015).
Eng, C.-H. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568, 235–239 (2019).
Xia, C., Fan, J., Emanuel, G., Hao, J. & Zhuang, X. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc. Natl Acad. Sci. USA 116, 19490–19499 (2019).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981 (2018).
Lin, J. R., Fallahi-Sichani, M. & Sorger, P. K. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 6, 8390 (2015).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Mercer, T. R. et al. The human mitochondrial transcriptome. Cell 146, 645–658 (2011).
Gudenas, B. L. & Wang, L. Prediction of LncRNA subcellular localization with deep learning from sequence features. Sci. Rep. 8, 16385 (2018).
Baker, S. C. et al. The External RNA Controls Consortium: a progress report. Nat. Methods 2, 731–734 (2005).
Ghandi, M. et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019).
Nusinow, D. P. et al. Quantitative proteomics of the Cancer Cell Line Encyclopedia. Cell 180, 387–402 (2020).
National Cancer Institute. NCI-60 Human Tumor Cell Lines Screen https://dtp.cancer.gov/discovery_development/nci-60/ (2020).
Kinker, G. S. et al. Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat. Genet. 52, 1208–1218 (2020).
Liu, J. et al. Concordance of MERFISH spatial transcriptomics with bulk and single-cell RNA sequencing. Preprint at bioRxiv https://doi.org/10.1101/2022.03.04.483068 (2022).
Leader, A. M. et al. Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification. Cancer Cell 39, 1594–1609 (2021).
Danaher, P. et al. Advances in mixed cell deconvolution enable quantification of cell types in spatially-resolved gene expression data. Nat. Commun. 13, 385 (2022).
Illumina. Evaluating RNA quality from FFPE samples. https://www.illumina.com/content/dam/illumina-marketing/documents/products/technotes/evaluating-rna-quality-from-ffpe-samples-technical-note-470-2014-001.pdf (2021).
Leica Biosystems. OND-III fully automated IHC and ISH staining system. https://www.leicabiosystems.com/ihc-ish-fish/fully-automated-ihc-ish-instruments/bond-iii/ (2021).
Qiu, P. Embracing the dropouts in single-cell RNA-seq analysis. Nat. Commun. 11, 1169 (2020).
Lin, J. R., Fallahi-Sichani, M., Chen, J. Y. & Sorger, P. K. Cyclic immunofluorescence (CycIF), a highly multiplexed method for single-cell imaging. Curr. Protoc. Chem. Biol. 8, 251–264 (2016).
Caruthers, M. H. et al. Chemical synthesis of deoxyoligonucleotides by the phosphoramidite method. Methods Enzymol. 154, 287–313 (1987).
Zhang, Z., Revyakin, A., Grimm, J. B., Lavis, L. D. & Tjian, R. Single-molecule tracking of the transcription cycle by sub-second RNA detection. eLife 3, e01775 (2014).
Wagle, M.-C. et al. A transcriptional MAPK Pathway Activity Score (MPAS) is a clinically relevant biomarker in multiple cancer types. NPJ Precis. Oncol. 2, 7 (2018).
Son, Y. H. et al. Roles of MAPK and NF-kappaB in interleukin-6 induction by lipopolysaccharide in vascular smooth muscle cells. J. Cardiovasc. Pharmacol. 51, 71–77 (2008).
Kang, H. B., Kim, Y. E., Kwon, H. J., Sok, D. E. & Lee, Y. Enhancement of NF-kappaB expression and activity upon differentiation of human embryonic stem cell line SNUhES3. Stem Cells Dev. 16, 615–623 (2007).
Hiscott, J. et al. Induction of human interferon gene expression is associated with a nuclear factor that interacts with the NF-kappa B site of the human immunodeficiency virus enhancer. J. Virol. 63, 2557–2566 (1989).
Kitamura, A., Takahashi, K., Okajima, A. & Kitamura, N. Induction of the human gene for p44, a hepatitis-C-associated microtubular aggregate protein, by interferon-alpha/beta. Eur. J. Biochem. 224, 877–883 (1994).
Kim, J. H., Park, S. Y., Jun, Y., Kim, J. Y. & Nam, J. S. Roles of Wnt target genes in the journey of cancer stem cells. Int. J. Mol. Sci. 18, 1604 (2017).
Jho, E. H. et al. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22, 1172–1183 (2002).
Lustig, B. et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 22, 1184–1193 (2002).
Yan, D. et al. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta–catenin signaling is activated in human colon tumors. Proc. Natl Acad. Sci. USA 98, 14973–14978 (2001).
Ramakrishnan, A. B. & Cadigan, K. M. Wnt target genes and where to find them. F1000Res 6, 746 (2017).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
He, T. C. et al. Identification of c-MYC as a target of the APC pathway. Science 281, 1509–1512 (1998).
Shtutman, M. et al. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl Acad. Sci. USA 96, 5522–5527 (1999).
Katoh, Y. & Katoh, M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr. Mol. Med. 9, 873–886 (2009).
Danaher, P. et al. Gene expression markers of Tumor Infiltrating Leukocytes. J. Immunother. Cancer 5, 18 (2017).
Nguyen, Q. H. et al. Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity. Nat. Commun. 9, 2028 (2018).
Barneda, D. et al. The brown adipocyte protein CIDEA promotes lipid droplet fusion via a phosphatidic acid-binding amphipathic helix. eLife 4, e07485 (2015).
Ussar, S. et al. ASC-1, PAT2, and P2RX5 are cell surface markers for white, beige, and brown adipocytes. Sci. Transl. Med. 6, 247ra103 (2014).
Min, S. Y. et al. Diverse repertoire of human adipocyte subtypes develops from transcriptionally distinct mesenchymal progenitor cells. Proc. Natl Acad. Sci. USA 116, 17970–17979 (2019).
Shan, T., Liu, W. & Kuang, S. Fatty acid binding protein 4 expression marks a population of adipocyte progenitors in white and brown adipose tissues. FASEB J. 27, 277–287 (2013).
Bradley, D. & Roth, G. Adaptive thresholding using the integral image. J. Graph. Tools 12, 13–21 (2007).
Krishnamoorthy, A. & Menon, D. Matrix inversion using Cholesky decomposition. In 2013 Signal Processing: Algorithms, Architectures, Arrangements, and Applications (SPA) (IEEE, 2013).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Acknowledgements
Research and development reported in this publication was supported in part through a strategic development collaboration between NanoString Technologies and Lam Research (Fremont, CA). The authors thank B. Birditt, B. Filanoski, J. Jenkins and E. Zhao from NanoString Technologies for provision of technical support.
Author information
Authors and Affiliations
Contributions
S.H. undertook conception and design of the work, supervised data collection, analysis and interpretation and drafted the manuscript. R.B. developed protein data processing algorithms. C.B. was responsible for instrument control software, SMI automation and integration of all instrument subsystems software. E.A.B. reviewed and validated protein data (Fig. 6) and performed the nested protein multiplex validation experiment (Supplementary Fig. 17). D.L.B. developed manual and automated processes for inventorying, quantitation, normalization, pooling, purification and quality control of oligonucleotides in 980-plex RNA panel and SMI reporters. K.C. performed oligo conjugations of antibodies used in SMI protein assays. P.D. designed the 980-plex RNA panel with E.P., performed comparison with RNA-seq in Fig. 2, performed NSCLC analyses in Fig. 3, performed reproducibility analysis in Fig. 5 and contributed to manuscript writing/editing. D.D. led the team that developed the system, instrumentation and software. R.G.G. developed reporter chemistry, assembly, quality control and manufacturing, and contributed to the section SMI reporter design and assembly and Supplementary Fig. 18. G.G. led development of protein-based SMI assays and design and interpretation of protein experiments. M.T.G. analyzed profiling data of human lung cells. M.L.H. contributed to the development of reporters and analysis of FFPE RNA quality and created Supplementary Fig. 6. R.K. designed and developed reporter structure. E.E.K. contributed to SMI methods. D.K. developed the overall concept, designed and guided experiments and analyzed data. T.K.K. supervised data collection and interpretation of human lung samples. Y.K. supervised data collection, analysis and interpretation of human lung samples. A. Klock developed the SMI ISH probe design pipeline and designed SMI ISH probes used in the 980-plex RNA panel. M.K. developed the primary data analysis pipeline for RNA and protein targets. A. Kutchma designed SMI ISH probes and wrote the section SMI ISH probe design. Z.R.L. designed and developed the protein assay, executed protein analyses and performed manuscript writing and editing. Y.L. conducted a pathological review to identify the correct staining pattern for all antibodies used. J.S.N.’s team was responsible for all chemistry process development efforts and supply chain management pertaining to the outsourcing of oligonucleotide synthesis, process improvements, price negotiation and supply agreement, key component scale-up and validation of all custom reagents and DNA components in R&D required for SMI—including contract manufacturing operation and validation of the PC spacer, synthesis and quality control of the three large-scale component sets needed for SMI reporter manufacturing plus high-throughput synthesis, outsourcing, quality control and processing of the thousands of required ISH probes. G.T.O. developed morphology and segmentation markers. E.P.P. developed the SMI instrument optical subsystem, instrument validation and support. J.C.P. developed the encoding scheme, screened reporter sequences and developed readout sequences. T.P.-E. optimized protein assay and undertook data collection, protein analyses and manuscript writing/editing. E.P. designed the 980-plex RNA panel with P.D. and contributed to manuscript writing/editing. T.R. developed the secondary analysis and target decoding pipeline for RNA targets, and the SMI instrument fluidic subsystem and workflow software. Z.R. developed the LR interaction analysis method, analyzed LR interactions across all tumors and contributed to manuscript writing/editing. M.R. performed initial chemistry development, supervised data release and contributed to writing the manuscript. A.R. was responsible for SMI protein assay content design and reagent validation. D.R. created plots of transcript positions overlaid on morphology and segmentation images for the figures. H.S. carried out manuscript development, writing and editing and created figures and tables. A.W.W. designed and developed the cell segmentation pipeline and contributed to manuscript writing/editing. C.A.W.-W. performed lung RNA isolation, processing and quality measurement and NGS library preparation and sequencing. L.W. established the cell segmentation pipeline, optimized the on-instrument SMI readout workflow and contributed to manuscript writing/editing. J.M.B. conceived the project, helped with experimental design and analysis and contributed to manuscript writing/editing.
Corresponding author
Ethics declarations
Competing interests
All authors are employees of NanoString Technologies and hold NanoString stock or stock options. D.K. is an employee of Dxome Co.
Peer review
Peer review information
Nature Biotechnology thanks Sanjay Tyagi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–18.
Supplementary Tables 1–11.
Supplementary Tables 1–11; combined tables separated by tab.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, S., Bhatt, R., Brown, C. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol 40, 1794–1806 (2022). https://doi.org/10.1038/s41587-022-01483-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-022-01483-z
This article is cited by
-
Spatial multi-omics: novel tools to study the complexity of cardiovascular diseases
Genome Medicine (2024)
-
Library size confounds biology in spatial transcriptomics data
Genome Biology (2024)
-
scGIST: gene panel design for spatial transcriptomics with prioritized gene sets
Genome Biology (2024)
-
Unsupervised spatially embedded deep representation of spatial transcriptomics
Genome Medicine (2024)
-
Charting multicellular tissue structure cell-to-cell
Nature Genetics (2024)