Abstract
Transplantation of B cells engineered ex vivo to secrete broadly neutralizing antibodies (bNAbs) has shown efficacy in disease models. However, clinical translation of this approach would require specialized medical centers, technically demanding protocols and major histocompatibility complex compatibility of donor cells and recipients. Here we report in vivo B cell engineering using two adeno-associated viral vectors, with one coding for Staphylococcus aureus Cas9 (saCas9) and the other for 3BNC117, an anti-HIV bNAb. After intravenously injecting the vectors into mice, we observe successful editing of B cells leading to memory retention and bNAb secretion at neutralizing titers of up to 6.8 µg ml−1. We observed minimal clustered regularly interspaced palindromic repeats (CRISPR)–Cas9 off-target cleavage as detected by unbiased CHANGE-sequencing analysis, whereas on-target cleavage in undesired tissues is reduced by expressing saCas9 from a B cell-specific promoter. In vivo B cell engineering to express therapeutic antibodies is a safe, potent and scalable method, which may be applicable not only to infectious diseases but also in the treatment of noncommunicable conditions, such as cancer and autoimmune disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data are available in the main text, in the Extended Data figures and Supplementary Data and materials. Illumina sequencing data can be accessed in the SRA database under accession code PRJNA706552. Source data are provided with this paper.
References
Mendoza, P. et al. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 561, 479–484 (2018).
Bar-On, Y. et al. Safety and antiviral activity of combination HIV-1 broadly neutralizing antibodies in viremic individuals. Nat. Med. 24, 1701–1707 (2018).
Cohen, Y. Z. et al. Safety, pharmacokinetics, and immunogenicity of the combination of the broadly neutralizing anti-HIV-1 antibodies 3BNC117 and 10-1074 in healthy adults: a randomized, phase 1 study. PLoS ONE 14, e0219142 (2019).
Johnson, P. R. et al. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. 15, 901–906 (2009).
Balazs, A. B. et al. Vectored immunoprophylaxis protects humanized mice from mucosal HIV transmission. Nat. Med. 20, 296–300 (2014).
Priddy, F. H. et al. Adeno-associated virus vectored immunoprophylaxis to prevent HIV in healthy adults: a phase 1 randomised controlled trial. Lancet HIV 6, e230–e239 (2019).
Nahmad, A. D. et al. Engineered B cells expressing an anti-HIV antibody enable memory retention, isotype switching and clonal expansion. Nat. Commun. 11, 5851 (2020).
Huang, D. et al. Vaccine elicitation of HIV broadly neutralizing antibodies from engineered B cells. Nat. Commun. 11, 5850 (2020).
Fusil, F. et al. A lentiviral vector allowing physiologically regulated membrane-anchored and secreted antibody expression depending on B-cell maturation status. Mol. Ther. 23, 1734–1747 (2015).
Greiner, V. et al. CRISPR-mediated editing of the B cell receptor in primary human B cells. Science 12, 369–378 (2019).
Voss, J. E. et al. Reprogramming the antigen specificity of B cells using genome-editing technologies. eLife 8, e42995 (2019).
Hartweger, H. et al. HIV-specific humoral immune responses by CRISPR/Cas9-edited B cells. J. Exp. Med. 216, 1301–1310 (2019).
Moffett, H. F. et al. B cells engineered to express pathogen-specific antibodies protect against infection. Sci. Immunol. 4, eaax0644 (2019).
Smith, T. T. et al. In situ programming of leukaemia-specific t cells using synthetic DNA nanocarriers. Nat. Nanotechnol. 12, 813–822 (2017).
Agarwal, S. et al. In vivo generation of CAR T cells selectively in human CD4+ lymphocytes. Mol. Ther. 28, 1783–1794 (2020).
Agarwal, S., Weidner, T., Thalheimer, F. B. & Buchholz, C. J. In vivo generated human CAR T cells eradicate tumor cells. Oncoimmunology 8, e167161 (2019).
Frank, A. M. et al. Combining T-cell-specific activation and in vivo gene delivery through CD3-targeted lentiviral vectors. Blood Adv. 4, 5702–5715 (2020).
Frank, A. M. & Buchholz, C. J. Surface-engineered lentiviral vectors for selective gene transfer into subtypes of lymphocytes. Mol. Ther. Methods Clin. Dev. 12, 19–31 (2019).
Frank, A. M. et al. CD8-specific designed ankyrin repeat proteins improve selective gene delivery into human and primate T lymphocytes. Hum. Gene Ther. 31, 679–691 (2020).
Pfeiffer, A. et al. In vivo generation of human CD19-CAR T cells results in B-cell depletion and signs of cytokine release syndrome. EMBO Mol. Med. 10, e9158 (2018).
Michels, A. et al. Lentiviral and adeno-associated vectors efficiently transduce mouse T lymphocytes when targeted to murine CD8. Mol. Ther. Methods Clin. Dev. 23, 334–347 (2021).
Münch, R. C. et al. Off-target-free gene delivery by affinity-purified receptor-targeted viral vectors. Nat. Commun. 6, 2–10 (2015).
Breuer, C. B. et al. In vivo engineering of lymphocytes after systemic exosome-associated AAV delivery. Sci. Rep. 10, 4544 (2020).
Nawaz, W. et al. AAV-mediated in vivo CAR gene therapy for targeting human T-cell leukemia. Blood Cancer J. 11, 119 (2021).
Rurik, J. G. et al. CAR T cells produced in vivo to treat cardiac injury. Science 375, 91–96 (2022).
Parayath, N. N., Stephan, S. B., Koehne, A. L., Nelson, P. S. & Stephan, M. T. In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells in vivo. Nat. Commun. 11, 6080 (2020).
Grimm, D. et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 82, 5887–5911 (2008).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Scheid, J. F. et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011).
Hung, K. L. et al. Engineering protein-secreting plasma cells by homology-directed repair in primary human B cells. Mol. Ther. 26, 456–467 (2018).
Mesin, L., Ersching, J. & Victora, G. D. Germinal center B cell dynamics. Immunity 45, 471–482 (2016).
Sok, D. & Burton, D. R. HIV broadly neutralizing antibodies: taking good care of the 98%. Immunity 45, 958–960 (2016).
Garber, D. A. et al. Durable protection against repeated penile exposures to simian-human immunodeficiency virus by broadly neutralizing antibodies. Nat. Commun. 11, 3195 (2020).
Taylor, J. J., Pape, K. A., Steach, H. R. & Jenkins, M. K. Apoptosis and antigen affinity limit effector cell differentiation of a single naïve B cell. Science 347, 11214–11218 (2015).
Abbott, R. K. et al. Precursor frequency and affinity determine B cell competitive fitness in germinal centers, tested with germline-targeting HIV vaccine immunogens. Immunity 48, 133–145.e6 (2017).
Dosenovic, P. et al. Anti-HIV-1 B cell responses are dependent on B cell precursor frequency and antigen binding affinity. Proc. Natl Acad. Sci. USA 115, 4743–4748 (2018).
Dey, B. et al. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 5, e1000445 (2009).
Steichen, J. M. et al. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity 45, 483–496 (2016).
Lecomte, E. et al. Advanced characterization of DNA molecules in rAAV vector preparations by single-stranded virus next-generation sequencing. Mol. Ther. Nucleic Acids 4, e260 (2015).
Lazzarotto, C. R. et al. CHANGE-seq reveals genetic and epigenetic effects on CRISPR–Cas9 genome-wide activity. Nat. Biotechnol. 38, 1317–1327 (2020).
Busch, S. et al. Circulating monocytes and tumor‐associated macrophages express recombined immunoglobulins in glioblastoma patients. Clin. Transl. Med. 8, 18 (2019).
Gong, X. et al. Macrophage-derived immunoglobulin M inhibits inflammatory responses via modulating endoplasmic reticulum stress. Cells 10, 2812 (2021).
Fuchs, T. et al. Expression of combinatorial immunoglobulins in macrophages in the tumor microenvironment. PLoS ONE 13, e0204108 (2018).
Moreau, T., Bardin, F., Imbert, J., Chabannon, C. & Tonnelle, C. Restriction of transgene expression to the B-lymphoid progeny of human lentivirally transduced CD34+ cells. Mol. Ther. 10, 45–56 (2004).
McCarron, M. J., Park, P. W. & Fooksman, D. R. CD138 mediates selection of mature plasma cells by regulating their survival. Blood 129, 2749–2759 (2017).
Nemazee, D. Mechanisms of central tolerance for B cells. Nat. Rev. Immunol. 17, 281–294 (2017).
Russell, D. M. et al. Peripheral deletion of self-reactive B cells. Nature 354, 308–311 (1991).
Edraki, A., Mir, A., Ibraheim, R., Gainetdinov, I. & Sontheimer, E. J. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol. Cell 73, 714–726 (2019).
Pausch, P. et al. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 337, 333–337 (2020).
Hartmann, J. et al. A library-based screening strategy for the identification of DARPins as ligands for receptor-targeted AAV and lentiviral vectors. Mol. Ther. Methods Clin. Dev. 10, 128–143 (2018).
Barzel, A. et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature 517, 360–364 (2015).
Magoč, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Lee, H., Chang, H. Y., Cho, S. W. & Ji, H. P. CRISPRpic: fast and precise analysis for CRISPR-induced mutations via prefixed index counting. NAR Genom. Bioinform. 2, lqaa012 (2020).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Stern, A. et al. Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 35, 506–511 (2007).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995).
Wieland, A. et al. Defining HPV-specific B cell responses in patients with head and neck cancer. Nature 597, 274–278 (2020).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the \(2^{-{{\Delta}{\Delta}}_{C_{\mathrm{T}}}}\) method. Methods 25, 402–408 (2001).
Nojima, T. et al. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat. Commun. 2, 411–465 (2011).
Ferrari, S. et al. Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking. Nat. Biotechnol. 38, 1298–1308 (2020).
Santana-Magal, N. et al. Melanoma-secreted lysosomes trigger monocyte-derived dendritic cell apoptosis and limit cancer immunotherapy. Cancer Res. 80, 1942–1956 (2020).
Brinkman, E. K., Chen, T., Amendola, M. & Van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
Bitton, A., Nahary, L. & Benhar, I. in Phage Display: Methods and Protocols (eds. Hust, M. & Lim, T. S.) 349–363 (Springer, 2018).
Acknowledgements
We thank the Veterinary Service Center, Tel Aviv University for animal husbandry. The IDRFU, Genomic Research Unit and SICF units, Tel Aviv University for logistic support and council. We also thank L. Vardi, M. Gelbart, H. Kobo, D. Burstein, I. Benhar, N. Freund, M. Kay, T. Akriv and N. Gritsenko for reagents and feedback. This research was funded by the Varda and Boaz Dotan donation (A.B.), the H2020 European Research Council grant no. 759296 570 (A.B.) and the Israel Science Foundation grant nos. 1632/16, 2157/16 and 2876/21 (A.B.), The Bill and Melinda Gates Foundation grant no. OPP1183956 (J.E.V.), National Institutes of Health grant nos. R01 AI167003-01 (A.B.) AI128836 and R01 AI073148 (D.N.), Edmond J. Safra Center for Bioinformatics fellowship (T.K. and A.S.), St. Jude Children’s Research Hospital and ALSAC, National Institutes of Health Office Of The Director, Somatic Cell Genome Editing initiative grant no. U01AI157189 (S.Q.T.), the Gertner Institute Scholarship, the Yoran Institute Scholarship and the SAIA Foundation (A.D.N.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
A.D.N. designed, performed and analyzed the study. C.R.L. performed CHANGE-seq; S.Q.T. supervised CHANGE-seq experiments. N.Z. and T.K. performed bioinformatical analyses. A.S. and R.R.-A. supervised the bioinformatical analyses. N.Z., M.H.-F. and I.R. helped with sample processing. Y.R. helped with vector design and cloning. D. Nataf and I.D. designed the B cell progenitor enrichment. M.T. and D.H. performed neutralization assays. D. Nemazee and J.E.V. supervised neutralization assays. I.D. contributed to supervising the study. Y.C. helped with experimental design. A.D.N. and A.B. drafted and revised the manuscript. A.B. conceptualized and supervised the study.
Corresponding author
Ethics declarations
Competing interests
A.D.N., D. Nataf, M.H.-F., I.D. and A.B. are listed as inventors on patent applications covering B cell engineering. A.D.N. and A.B. have an equity stake in and receive monetary compensation from Tabby Therapeutics Ltd, a B cell engineering company. S.Q.T. is a coinventor on patents covering the CHANGE-seq method. S.Q.T. is a member of the scientific advisory boards of Kromatid, Inc. and Twelve Bio. The other authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Multiple isotypes of the 3BNC117 antibody are expressed by engineered B cells.
a–d. ELISA for each isotype. a. IgM, b. IgG1, c. IgG2c and d. IgA. All samples come from the CMV-Cas9gRNA + donor injected mice at different time points, as indicated in each legend. Mean and SD are indicated. n = 3 biologically independent animals. e,f. Area under the curve (AUC) for A-D. e. IgM, ns; pv = 0.1288, ***; pv = 0.0008, **; pv = 0.0062, f. IgG1, ns; pv = 0.131 and from top to bottom as presented in the graph, **; pv = 0.0044 and pv = 0.0013, g. IgG2c, ***; pv = 0.0007, *; pv = 0.0195, **; pv = 0.0098, h. IgA, ns; pv = 0.0587, **; = pv = 0.0013, *; pv = 0.0403, for two-sided unpaired t-test. n = 3 biologically independent animals. For A-H, sample collection day is indicated. Mean values are indicated. i. Fraction of 3BNC117 IgG titers as quantified by ELISA using purified gp120 binding sera from donor injected mice immunized with gp120, at day 37. ***; pv = 0.0007 for two-sided unpaired t-test. n = 3 biologically independent samples. Mean values are indicated.
Extended Data Fig. 2 bNAb genomic integration, sera titers and neutralization as a function of immunizations and co-injection of the CRISPR-Cas9 vector.
a. Area under the Curve (AUC) of Fig. 2d for YU2.DG (left) and JRFL (right) **; pv = 0.0036 (YU2.DG) and pv = 0.005 (JRFL) for unpaired t-test for CMV-Cas9gRNA + donor to PBS comparison and ##; pv = 0.0072 (YU2.DG) and pv = 0.0063 (JRFL) for one-sample t-test for Naïve to PBS comparison. n = 3 for CMV-Cas9gRNA + donor and PBS. Naïve sample is from a single, non-immunized, non-AAV-injected mouse. Mean values ± SD are indicated. b. Area under the curve (AUC) of Fig. 2c. From top to bottom, *; pv = 0.0185 and pv = 0.0103, **; pv = 0.0036 for two-sided unpaired t-test. n = 3 biologically independent animals. Mean values ± SEM are indicated. c. RT-PCR on RNA from sorted, 3BNC117+, CD19+, CD4− blood lymphocytes from day 37. Here, we used a reverse primer in a membranal exon of either IgHCμ or IgHCγ (all subtypes) and a forward primer on the VH of the coded 3BNC117. Numbers indicate different mice, injected with either a) PBS, b) the donor vector and the CMV-Cas9gRNA vector, or c) the donor vector only, as indicated above the gels. Control sample (C+) comes from in-vitro engineered primary mouse splenic lymphocytes, as described previously7. Ladder sizes are indicated on the left. Arrow indicates the expected amplicon size. For each group, experiment was reproduced 3 times with independent samples, as indicated by the numbers. Molecular weight markers (M) and their respective size in base pairs (MW) are indicated. d. Total DNA from the previous reaction as in (C) was purified and a semi-nested PCR with the same forward primer and a reverse primer on the CH1 of the respective constant domains. Ladder sizes are indicated on the left. Arrow indicates the expected amplicon size. For each group, experiments were reproduced 3 times with independent samples. Molecular weight markers (M) and their respective size in base pairs (MW) are indicated. e. Sanger sequencing alignment and chromatogram of the purified amplicon from the previous step. Reference sequences are indicated above. For the IgHCγ, each subtype reference is indicated. Sequencing of the IgHCμ amplicon of donor 3 has failed. f. Experimental design for (G-I). Splenic lymphocytes were activated with LPS and IL-4 and engineered, ex vivo, by AAV transduction and Cas9 electroporation with or without a gRNA. h. Flow cytometry of engineered splenic lymphocytes two days following treatment. Pregated on live, singlets. FcR block was used in the staining. Engineering parameters are indicated above each plot. h. EtBr gel electrophoresis showing products of an RT-PCR reaction with RNA from cells two days following treatment as in (F). For each sample, a control (C) reaction was performed amplifying the endogenous IgHG1 cDNA. Ladder sizes are indicated on the left. Arrow indicates the expected amplicon size. The experiment was reproduced once, with similar results. Molecular weight markers (M) and their respective size in base pairs (MW) are indicated. i. Sanger sequencing of the previous amplicons, confirming the integration.
Extended Data Fig. 3 Detection of engineered B cells in the spleen, the blood and the bone marrow.
a. Flow cytometry plots demonstrating 3BNC117 expression among CD19+ CD11b− cells in the spleen at day 82 of 2CC immunized mice. Pregated on live, singlets. FcR block was used in the staining. b. Quantification of B. ***; pv = 0.0006 for two-sided unpaired t-test. c. Flow cytometry plots demonstrating 3BNC117 expression among blood B cells (CD19+, CD4−). d. Quantification of blood 3BNC117-expressing cells over time. The black arrows indicate immunizations and the blue arrow indicates AAV injection. ####; pv < 0.0001 for Two-Way ANOVA comparison between groups and *; pv = 0.0133, ****; pv < 0.0001 Two-Way ANOVA with Šidák’s multiple comparison for time points comparison to PBS. For each group, each line represents the mean ±SD of n = 3 biologically independent mice. e. Quantification of total CD38+ CD138+ in spleens of recipient mice as in Fig. 3a. From top to bottom: ns; pv = 0. 2380 and pv = 0.9907 for One-Way ANOVA with Tukey’s multiple comparisons test. f. Quantification of total GL7+ CD95+ cells as in Fig. 3e. From top to bottom: ns; pv = 0.9857 and pv = 0.9985 for One-Way ANOVA with Tukey’s multiple comparisons test. g. Flow cytometry plots demonstrating the presence of 3BNC117-expressing B cells in the bone marrow. h-i. Quantification of G. ns; pv = 0.9965, ****; pv < 0.0001 (G) and from top to bottom: ns; pv > 0.9999, *; pv = 0.0387 and pv = 0.0372 (H) for One-Way ANOVA with Tukey’s multiple comparison.
Extended Data Fig. 4 Assessing expression of the transgene in different subsets of cells.
a. Flow cytometry examples for Fig. 5c-g and Extended Data Fig. 3b-e, j-k. b. Quantification of 3BNC117+ cells in bone marrow. #; pv = 0.0129 for Two-Way ANOVA and *; pv = 0.0255 for Two-Way ANOVA with Tukey’s multiple comparison. c. Quantification of the indicated populations from 3BNC117+ B220− cells in the bone marrow. *; pv = 0.0335, **** = pv < 0.0001 for One-Way ANOVA with Tukey’s multiple comparison. d,e. Quantification of GFP+ cells from the CD11b+ 3BNC117+ or CD11b+ 3BNC117− populations in the bone marrow (D) or spleen (E). *; pv = 0.0214, **** = pv <0.0001 for unpaired two-tailed t-test. Mean values are indicated. f. Experimental scheme for (G). Bone marrow cells were collected and activated in the presence of GMCSF. 5 days later, cells were cultured in the presence of purified mouse 3BNC117 for FcR presentation and then collected for analysis by flow cytometry. g. Analysis by flow cytometry, using the anti-3BNC117 anti-idiotypic antibody, of bone marrow cells activated by GMCSF and subsequently cultured with 3BNC117. Experimental conditions are indicated above. Pre-gated on CD11b, live, singlets. h. Experimental scheme for (I). Splenic Lymphocytes are collected from mice and cultured with purified mouse 3BNC117. Next, cells are collected for Flow Cytometry. i. Analysis by flow cytometry, using the anti-3BNC117 anti-idiotypic antibody, of splenic lymphocytes cultured with 3BNC117. Experimental conditions are indicated above. Pre-gated on B220+, live, singlets. j-k. Quantification of 3BNC117+ cells from GFP+ cells in the spleen (J) ####; pv < 0.0001 for Two-Way ANOVA and ns; pv = 0.1748, ****; pv < 0.0001 for Two-Way ANOVA with Tukey’s multiple comparison or bone marrow (K) ###; pv = 0.001 for Two-Way ANOVA and ns; pv = 0.6177, and from left to righ: ***; pv = 0.0005 and pv = 0.0005 for Two-Way ANOVA with Tukey’s multiple comparison. BM = bone marrow. l. Experimental scheme of in-vitro engineering of primary GMCSF activated bone marrow cells for (M-N). m. EtBr gel electrophoresis showing the product of an RT-PCR reaction of RNA from activated bone marrow cells, six days following treatment as in (L). For each sample, two reactions were performed (N or E). N amplifies endogenous IgHCµ mRNA. E amplifies the transgene mRNA joined by splicing to the IgHCµ exons following engineering. Indicated labeling of the amplicons (E1, E2, N1 and N2) performed for reference in (N). The experiment was performed once with expected results. Molecular weight markers (M) and their respective size in base pairs (MW) are indicated. n. Sanger sequencing of the amplicons, confirming correct integration. Amplicons are annotated E1, E2, N1 and N2 as in (M). o. Flow cytometry example of engineered GMCSF activated primary bone marrow cells. Pre-gated on B220−, live, singlets. p. quantification of O. Control cells are cells transduced with the AAV and electroporated only with the spCas9, without the sgRNA (-gRNA). For A-P and Fig. 3, FcR block was used in the staining. For each group, n = 2 biologically independent samples.
Extended Data Fig. 5 Engineering B cells with the sgRNA coded on the donor AAV.
a. Vector maps of the AAVs coding for the donorgRNA and the SFFV-Cas9. b. 3BNC117 IgG titers as quantified by ELISA over time in the SFFV-Cas9 + donorgRNA group. The black arrows indicate immunizations and the blue arrow indicates AAV injection. Each line represents a mouse. ***; pv = 0.0005 for Two-Way ANOVA comparing the SFFV-Cas9 + donorgRNA group to the donor group. n = 3 biologically independent mice. In this panel, the PBS and donor control groups are the same as for Fig. 2B. c. Transduction neutralization of TZM.bl cells by the YU2.DG (top) and JRFL (bottom) HIV pseudoviruses in the presence of day 136 sera IgGs. Neutralization is calculated as percent reduction from maximal luminescence per sample. The PBS control received immunizations, while the naïve control represents serum IgG from an untreated mouse. ns = non-significant, *; pv = 0.0462 and from top to bottom: ***; pv = 0.0007 and pv = 0.0004, ****; pv < 0.0001, Two-Way ANOVA with Šidák’s multiple comparison for time points comparison to PBS. d. Area under the Curve (AUC) for C. for YU2.DG and JRFL. ns; pv = 0.0667 (YU2.DG) and *; pv = 0.0103 (JRFL) for two-sided unpaired t-test for CMV-Cas9gRNA + donor to PBS comparison and ##; pv = 0.0097 (YU2.DG) and pv = 0.0078 (JRFL) for two-sided one-sample t-test for naïve to PBS comparison. n = 3 for CMV-Cas9gRNA + donor and PBS. Naïve sample from a single, non-immunized, non-AAV-injected mouse. In C-D, the PBS and control group is the same as for Fig. 2c and Extended Data Fig. 2a. Mean values ± SD are indicated. e. Representative flow cytometry analysis of 3BNC117+, CD19+, CD4− blood lymphocytes over time in the SFFV-Cas9 + donorgRNA group. f. Quantification of E. The black arrows indicate immunizations and the blue arrow indicates AAV injection. ###; pv = 0.0006 for Two-Way ANOVA comparison between groups and ****; pv < 0.0001 for Two-Way ANOVA with Šidák’s multiple comparison for time points comparison to PBS. For each group, each line represents the mean ±SD of n = 3 biologically independent mice. g. Representative flow cytometry analysis of 3BNC117+, CD19+, CD38+, CD138+ plasmablasts in the spleens of the SFFV-Cas9 + donorgRNA group at day 136. h. Quantification of G. Mean is indicated by the bars. ns; pv = 9892, ***; pv = 0.0005, one-way ANOVA with Tukey’s multiple comparison. i. Representative flow cytometry analysis of GL7+, Fas/CD95+ GC B cells in the spleens of the SFFV-Cas9 + donorgRNA group at day 136. j. Quantification of I. Mean is indicated by the bars. ns; pv = 0.8916, **; pv = 0.0075, one-way ANOVA with Tukey’s multiple comparison. k. Representative flow cytometry analysis of 3BNC117+ cells in total bone marrow (BM) of the SFFV-Cas9 + donorgRNA group at day 136. l. Quantification of 3BNC117+ CD19+ cells, ns; pv = 0.9965 and ****; pv < 0.0001 and m. Quantification of 3BNC117+ CD19− cells, ns; pv > 0.9999 and from top to bottom: ***; pv = 0.0003 and pv = 0.0003. Mean is indicated by the bars. One-way ANOVA with Tukey’s multiple comparison. n-o. Assessing overall immune homeostasis. Quantification by flow cytometry of total CD38+ CD138+ plasmablasts in spleen (N) ns; pv = 0.5622, total GL7+, Fas+ GC B cells in the spleen (O), at day 136 ns; pv = 0.9926. Mean is indicated by the bars.One-way ANOVA with Tukey’s multiple comparison. For E-O, the PBS and donor control groups are the same as for Fig. 3 and Extended Data Fig. 3.
Extended Data Fig. 6 Long-term persistence of serum antibodies and antibody-secreting cells in the bone marrow.
a. Experimental design. Here, all mice were immunized 4 times (days 8,23,38 and 53) and selected groups received 2 additional immunizations on days 68 and 83. b. 3BNC117 IgG titers as quantified by ELISA over time. For each group, each line represents the mean ±SD between day 22 and day 67 represents n = 6 for biologically independent mice and n = 3 for day 67 to 97. c-d. Representative ELISPOT experiments (C) and quantification (D) of 3BNC117-secreting cells from the bone marrow of mice of the indicated groups. Numbers in parentheses represent the number of immunizations. From top to bottom followed by left to right, ns; pv = 0.9844 and pv = 0.9963 and pv = 0.0884, *; pv = 0.0135, **; pv = 0.008, ***; pv = 0.0005 and pv = 0.0003 for One-Way ANOVA with Tukey’s multiple comparison. Mean values are indicated. Part of the data presented in panels B-D is presented also in Fig. 6c–e.
Extended Data Fig. 7 Using CD19 rather than CMV promoter, to drive saCas9 expression, reduces the engineering rate of B cell progenitors.
a. Origin of cells used in this experiment. Bone marrows were extracted 100 days following AAV injection and enriched for IL7R+. b. Experimental scheme. Enriched IL7R+ cells were grown in the presence of multiple activation factors as indicated in the representative timeline, numbers indicate days. Horizontal bars below the timeline indicate the presence of a specific factor supplemented to the growth media. c. Representative flow cytometry analysis of in−vitro differentiation of the IL7R+ enriched cells over time. Days as in (B) are indicated above the plots. d. Quantification of C. For each group, each line represents n = 3 biologically independent samples. e. Representative flow cytometry analysis of IL7R enrichment from mice, as in (A). f. Quantification of E. g. Representative flow cytometry analysis of 3BNC117+ CD19+ expression by cells, following 9 days of in−vitro differentiation, as in (B). h. Quantification of G. Each dot represents cells collected from a single mouse as in (A). ns; pv = 0.9, *; pv = 0.0122 and **; pv = 0.0077 for One-Way ANOVA with Tukey’s multiple comparison. For C-G, FcR block was used in staining.
Extended Data Fig. 8 Low 3BNC117 staining and ELISA levels are obtained following syngeneic transplantation of Lin- enriched cells from mice injected with the donor vector with or without the saCas9 coding vector.
a. Experimental scheme. CD45.2 mice received either a donor AAV expressing the gRNA or both a donor AAV and an AAV expressing saCas9 and the gRNA. Immunization protocol is indicated by the bars in black. On day 97, bone marrow cells were collected, enriched for lineage negative cells (Lin-) and transplanted into, recipient CD45.1 mice. Recipient mice are sublethally irradiated before transplantation and immunized after transplantation. b-c. Representative flow cytometry analysis (B) and quantification (C) of the enriched Lin- population from the donor mice. ns; pv = 0.9627 for One-Way ANOVA with Tukey’s multiple comparison. d. Representative flow cytometry analysis (D) of spleens (top) or bone marrows (bottom) from recipient mice at day 140, as in (A) e. Quantification of D. for the CD45.2+ CD45.1- population. From top to bottom: ns; pv = 0.4595 and pv > 0.9999 for Two-Way ANOVA with Tukey’s multiple comparison. f-g. Quantification of D. for the rate of 3BNC117-expressing cells for the spleen (F) or the bone marrow (G). From top to bottom: ns; pv = 0.9994 and pv = 0.0668 (F) and pv = 0.2371 and pv = 0.0560 for Two-Way ANOVA with Tukey’s multiple comparison. h. Serum 3BNC117 IgG titers at the indicated time points. The scale of the Y-axis was chosen to correspond to the other 3BNC117 titer plots in this manuscript. ns; pv = 0.1726t for Two-Way ANOVA. i. Representative ELISPOT assay of a day 140 bone marrow from CD45.1 recipient mice. j. Quantification I. ns; pv = 0.1756 for two-sided unpaired t-test. For B-D, FcR block was used in staining.
Extended Data Fig. 9 Gating strategy for each experiment in this study.
Gating strategy for each experiment in this study.
Extended Data Fig. 10 Assessment of the specificity and sensitivity of the anti-idiotype for 3BNC117.
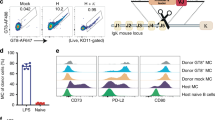
a. Experimental scheme. Cells are engineered a day following extraction from spleen and activation. We used three different donor AAVs, each expressing a different antibody: either 3BNC117, VRC01 or 10−1074. b. Flow cytometry of gp120 or anti-idiotype binding of engineered cells, two days following treatment. Staining procedure is indicated above the plots. FcR block was used in staining. Each row indicates a different AAV used. Untransduced cells serve as the negative control. c. Quantification of B. ****; pv < 0.0001 for Two-Way ANOVA with Tukey’s multiple comparison.
Supplementary information
Supplementary Table 1
Supplementary Table 1.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data and unprocessed gels.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data and unprocessed gels.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Nahmad, A.D., Lazzarotto, C.R., Zelikson, N. et al. In vivo engineered B cells secrete high titers of broadly neutralizing anti-HIV antibodies in mice. Nat Biotechnol 40, 1241–1249 (2022). https://doi.org/10.1038/s41587-022-01328-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-022-01328-9
This article is cited by
-
In vivo affinity maturation of mouse B cells reprogrammed to express human antibodies
Nature Biomedical Engineering (2024)
-
Identification of a novel role for TL1A/DR3 deficiency in acute respiratory distress syndrome that exacerbates alveolar epithelial disruption
Respiratory Research (2023)
-
Circular RNA vaccine in disease prevention and treatment
Signal Transduction and Targeted Therapy (2023)
-
B cell engineering in vivo: Accelerating induction of broadly neutralizing antibodies against HIV-1 infection
Signal Transduction and Targeted Therapy (2023)
-
CRISPR comes a-knock-in to reprogram antibodies in vivo
Nature Biotechnology (2022)