Abstract
Many cancers carry recurrent, change-of-function mutations affecting RNA splicing factors. Here, we describe a method to harness this abnormal splicing activity to drive splicing factor mutation-dependent gene expression to selectively eliminate tumor cells. We engineered synthetic introns that were efficiently spliced in cancer cells bearing SF3B1 mutations, but unspliced in otherwise isogenic wild-type cells, to yield mutation-dependent protein production. A massively parallel screen of 8,878 introns delineated ideal intronic size and mapped elements underlying mutation-dependent splicing. Synthetic introns enabled mutation-dependent expression of herpes simplex virus–thymidine kinase (HSV–TK) and subsequent ganciclovir (GCV)-mediated killing of SF3B1-mutant leukemia, breast cancer, uveal melanoma and pancreatic cancer cells in vitro, while leaving wild-type cells unaffected. Delivery of synthetic intron-containing HSV–TK constructs to leukemia, breast cancer and uveal melanoma cells and GCV treatment in vivo significantly suppressed the growth of these otherwise lethal xenografts and improved mouse host survival. Synthetic introns provide a means to exploit tumor-specific changes in RNA splicing for cancer gene therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
RNA-seq data from 16 normal human tissues (Illumina Body Map 2.0, illustrated in Fig. 1a) were downloaded from EMBL-EBI ArrayExpress (accession no. E-MTAB-513). RNA-seq data from published studies were downloaded from CGHub (TCGA cohorts), the Genomic Data Commons (accession BEATAML1.0-COHORT for the Beat AML cohort43), the Gene Expression Omnibus (accession no. GSE72790 for chronic lymphocytic leukemia22, GSE49642 for AML57, GSE63569 and GSE85712 for MDS15,42) and dbGaP (MDS58), or obtained directly from the authors (uveal melanoma10). High-throughput sequencing data generated as part of this study were deposited in the Gene Expression Omnibus (GEO accession GSE163217). All RNA-seq data was analyzed using the GRCh37/hg19 human genome assembly (GCF_000001405.13), as described above. All other data that support this study’s findings are available as Source Data for Figs. 1–6, Extended Data Figs. 1–6 and in Supplementary Tables provided with this paper.
Code availability
Software and algorithms used for analyzing alternative splicing in RNA-seq data, identifying SF3B1-mutant samples and mapping reads from the screens are published and described with citations in the relevant sections in Methods.
References
Yoshida, K. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011).
Papaemmanuil, E. et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. New Engl. J. Med. 365, 1384–1395 (2011).
Quesada, V. et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. https://doi.org/10.1038/ng.1032 (2011).
Graubert, T. A. et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. https://doi.org/10.1038/ng.1031 (2011).
Dvinge, H., Kim, E., Abdel-Wahab, O. & Bradley, R. K. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer 16, 413–430 (2016).
Wang, L. et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. New Engl. J. Med. 365, 2497–2506 (2011).
Harbour, J. W. et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. https://doi.org/10.1038/ng.2523 (2013).
Martin, M. et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. https://doi.org/10.1038/ng.2674 (2013).
Furney, S. J. et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Disc. https://doi.org/10.1158/2159-8290.cd-13-0330 (2013).
Alsafadi, S. et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 7, 10615 (2016).
Yoshimi, A. et al. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 574, 273–277 (2019).
The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature https://doi.org/10.1038/nature11412 (2012).
Mian, S. A. et al. SF3B1 mutant MDS-initiating cells may arise from the haematopoietic stem cell compartment. Nat. Commun. 6, 10004 (2015).
Lee, S. C.-W. et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat. Med. https://doi.org/10.1038/nm.4097 (2016).
Obeng, E. A. et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell 30, 404–417 (2016).
Shirai, C. L. et al. Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome. Nat. Commun. 8, 14060 (2017).
Wang, E. et al. Targeting an RNA-binding protein network in acute myeloid leukemia. Cancer Cell 35, 369–384.e7 (2019).
DeBoever, C. et al. Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLoS Comput. Biol. 11, e1004105 (2015).
Kim, E. et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell 27, 617–630 (2015).
Ilagan, J. O. et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 25, 14–26 (2015).
Brooks, A. N. et al. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS ONE 9, e87361 (2014).
Darman, R. B. et al. Cancer-associated SF3B1 hotspot mutations induce cryptic 3? Splice site selection through use of a different branch point. Cell Rep. 13, 1033–1045 (2015).
Zhang, J. et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1514105112 (2015).
Inoue, D. et al. Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature 574, 432–436 (2019).
Yeo, G. & Burge, C. B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 11, 377–394 (2004).
Gozani, O., Potashkin, J. & Reed, R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18, 4752–4760 (1998).
Spies, N., Nielsen, C. B., Padgett, R. A. & Burge, C. B. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell 36, 245–254 (2009).
Smith, K. O., Galloway, K. S., Kennell, W. L., Ogilvie, K. K. & Radatus, B. K. A new nucleoside analog, 9-[[2-hydroxy-1-(hydroxymethyl)ethoxyl]methyl]guanine, highly active in vitro against herpes simplex virus types 1 and 2. Antimicrob. Agents 22, 55–61 (1982).
Rosenberg, A. B., Patwardhan, R. P., Shendure, J. & Seelig, G. Learning the sequence determinants of alternative splicing from millions of random sequences. Cell 163, 698–711 (2015).
Wong, M. S., Kinney, J. B. & Krainer, A. R. Quantitative activity profile and context dependence of all human 5′ splice sites. Mol. Cell https://doi.org/10.1016/j.molcel.2018.07.033 (2018).
Liu, B. et al. Mutant SF3B1 promotes AKT and NF-kB driven mammary tumorigenesis. J. Clin. Invest. https://doi.org/10.1172/jci138315 (2020).
Dorer, D. E. & Nettelbeck, D. M. Targeting cancer by transcriptional control in cancer gene therapy and viral oncolysis. Adv Drug Deliver. Rev. 61, 554–571 (2009).
Lienert, F., Lohmueller, J. J., Garg, A. & Silver, P. A. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat. Rev. Mol. Cell Bio. 15, 95–107 (2014).
Wu, M.-R., Jusiak, B. & Lu, T. K. Engineering advanced cancer therapies with synthetic biology. Nat. Rev. Cancer 19, 187–195 (2019).
Culler, S. J., Hoff, K. G. & Smolke, C. D. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science 330, 1251–1255 (2010).
Watanabe, T. & Sullenger, B. A. Induction of wild-type p53 activity in human cancer cells by ribozymes that repair mutant p53 transcripts. Proc. Natl Acad. Sci. USA 97, 8490–8494 (2000).
Hong, S.-H. et al. In vivo reprogramming of hTERT by trans-splicing ribozyme to target tumor cells. Mol. Ther. 16, 74–80 (2008).
Kim, J. et al. Targeted regression of hepatocellular carcinoma by cancer-specific RNA replacement through microRNA regulation. Sci. Rep. 5, 12315 (2015).
Lee, S.-J. et al. Phase I trial of intravenous Ad5CRT in patients with liver metastasis of gastrointestinal cancers. Cancer Gene Ther. 26, 174–178 (2019).
Chung, H. K. et al. A compact synthetic pathway rewires cancer signaling to therapeutic effector release. Science 364, eaat6982 (2019).
Nissim, L. et al. Synthetic RNA-based immunomodulatory gene circuits for cancer immunotherapy. Cell 171, 1138–1150.e15 (2017).
Dolatshad, H. et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia https://doi.org/10.1038/leu.2014.331 (2014).
Tyner, J. W. et al. Functional genomic landscape of acute myeloid leukaemia. Nature 60, 277–531 (2018).
Pangallo, J. et al. Rare and private spliceosomal gene mutations drive partial, complete, and dual phenocopies of hotspot alterations. Blood https://doi.org/10.1182/blood.2019002894 (2020).
Lee, S. C.-W. et al. Synthetic lethal and convergent biological effects of cancer-associated spliceosomal gene mutations. Cancer Cell 34, 225–241.e8 (2018).
Griewank, K. G. et al. Genetic and molecular characterization of uveal melanoma cell lines. Pigment Cell Melanoma Res. 25, 182–187 (2012).
Pineda, J. M. B. & Bradley, R. K. Most human introns are recognized via multiple and tissue-specific branchpoints. Genes Dev. 32, 577–591 (2018).
Thomas, J. D. et al. RNA isoform screens uncover the essentiality and tumor-suppressor activity of ultraconserved poison exons. Nat. Genet. 52, 84–94 (2020).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Magoč, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Flicek, P. et al. Ensembl 2013. Nucleic Acids Res. 41, D48–D55 (2013).
Meyer, L. R. et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 41, D64–D69 (2013).
Katz, Y., Wang, E. T., Airoldi, E. M. & Burge, C. B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7, 1009–1015 (2010).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323 (2011).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Lavallée, V.-P. et al. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat. Genet. https://doi.org/10.1038/ng.3371 (2015).
Taylor, J. et al. Single-cell genomics reveals the genetic and molecular bases for escape from mutational epistasis in myeloid neoplasms. Blood 136, 1477–1486 (2020).
Acknowledgements
S.C. was supported by the American Society of Hematology and the Leukemia & Lymphoma Society. R.F.S. was supported in part by NIH/NCI (grant no. T32 CA009207). J.M.B.P. was supported in part by the ARCS Foundation. J.D.T. is a Washington Research Foundation Postdoctoral Fellow. J.T.P. was supported in part by NIH/NCI (grant no. T32 CA009657) and the Edward P. Evans Foundation. O.A.-W. and R.K.B. were supported in part by the Edward P. Evans Foundation, NIH/NCI (grant no. R01 CA251138), and NIH/NHLBI (grant no. R01 HL128239). O.A.-W. was supported in part by the NIH/NCI (grant nos. R01 CA242020 and P50 CA254838), The Leukemia & Lymphoma Society, and Henry and Marilyn Taub Foundation for MDS Research. R.K.B. was supported in part by the NIH/NIDDK (grant no. R01 DK103854) and NIH/NHLBI (grant no. R01 HL151651). R.K.B. is a Scholar of The Leukemia & Lymphoma Society (grant no. 1344-18) and holds the McIlwain Family Endowed Chair in Data Science. Computational studies were supported in part by FHCRC’s Scientific Computing Infrastructure (grant no. ORIP S10 OD028685). The results published here are based in part on data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Author information
Authors and Affiliations
Contributions
K.N., S.B., O.A.-W. and R.K.B. designed the study. K.N., S.B., B.L., S.C., R.F.S., J.P., J.M.B.P., J.T.P., A.E.B., A.M.G. and D.B.U. performed in vitro experiments. S.B, C.E. and H.C. performed in vivo experiments. M.S. and J.P.B. identified and provided patient materials. O.H. and H.-P.K. provided advice on experimental design. K.N. performed computational analyses. J.D.T. and J.T.P. assisted with experimental design and data interpretation. K.N., S.B., O.A.-W. and R.K.B. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
O.A.-W. has served as a consultant for H3B Biomedicine, Foundation Medicine Inc., Merck, Prelude Therapeutics and Janssen, and is on the Scientific Advisory Board of Envisagenics Inc., Pfizer Boulder and AIChemy Inc. O.A.-W. has received previous research funding from Loxo Oncology and H3B Biomedicine unrelated to the current paper. K.N., S.B., B.L., O.A.-W. and R.K.B. are inventors on a patent application submitted by Fred Hutchinson Cancer Research Center related to this work. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Validation of SF3B1 mutation-dependent differential splicing for endogenous and synthetic introns.
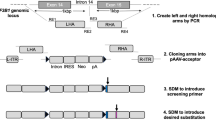
(a) As Fig. 1b, but additionally illustrating splicing patterns for normal bone marrow (n = 3) and cancer samples lacking SF3B1 mutations from each studied cohort. n = 3 randomly chosen SF3B1-WT samples illustrated for each cohort, with the exceptions of uveal melanoma, acute myeloid leukemia, and MDS, for which additional samples were illustrated given the high frequency of SF3B1 mutations in these disorders. (b) As Fig. 1b, but additionally including all samples with SF3B1K666E/N/R/T mutations with mutant allele expression > = 25%. (c) RT-PCR analysis of competing 3’ splice site (3’ss) usage within endogenous introns of ORAI2 and TMEM14C in K562 cells engineered to bear the indicated mutations in endogenous SF3B1. n = 4 biologically independent cell lines. (d) As (c), but for intron retention within endogenous introns of MYO15B and SYTL1. (e) RT-PCR analysis of endogenous MAP3K7 and MTERFD3 splicing in primary samples from patients with acute myeloid leukemia (AML, n = 8) as well as myelodysplastic syndromes (MDS, n = 9) and pancreatic ductal adenocarcinoma (PDAC) cell lines (n = 3) wild-type (WT) or mutant for SF3B1. The specific SF3B1 mutations present in each sample are indicated. Experiment repeated independently n = 3 times with similar results. (f) Sanger sequencing illustrating three distinct MTERFD3 isoforms arising from three competing 3’ss (two cryptic 3’ss and one canonical, frame-preserving, 3’ss). The three lower bands for the MTERFD3 RT-PCR illustrated in Fig. 1d were isolated, cloned, and sequenced to identify the specific 3’ss that were used for splicing of each isoform. (g) RT-PCR analysis of synthetic intron splicing for the indicated introns following transfection of the fluorescent reporter construct into isogenic K562 cells with the indicated SF3B1 genotypes. n = 2 biologically independent cell lines and n = 3 biologically independent experiments. (h) As (f), but for the indicated introns. n = 2 biologically independent cell lines. (i) Schematic of split HSV–TK construct with mCherry. This construct was used for the experiment illustrated in Fig. 2c. Uncropped gels are available as source data.
Extended Data Fig. 2 Hallmark SF3B1 mutation-responsive events are specific to SF3B1 mutations and recapitulated in breast epithelial cells.
(a) RNA-seq read coverage plot for K562 cells (top) and MCF10A cells (bottom) engineered to have the illustrated genotypes, illustrating specificity of mutant SF3B1-dependent usage of an intron-proximal cryptic 3’ss in MAP3K7. Each indicated mutant allele is present as a single copy in the endogenous locus in otherwise WT cells. Neither SRSF2 nor U2AF1 mutations induce the splicing changes caused by SF3B1 mutations. These RNA-seq data complement the related RT-PCR studies in Fig. 1d. (b) As (a), but for mutant SF3B1-dependent mis-splicing in MTERFD3. The MTERFD3 intron contains two specific splicing changes in SF3B1-mutant cells: increased intron excision (left) and increased usage of an intron-distal competing 3’ss (right). (c) Top, RT-PCR demonstrating mutation-dependent excision of the indicated synthetic introns in T47D cells expressing inducible SF3B1WT or SF3B1K700E (top). Bottom, relative viability of cells from top panel following treatment with ganciclovir. Data represented as mean ± s.d. n = 3 biologically independent experiments. (d) As (c), but for MOLM-13 cells. (e) As (c), but for Panc-1 (SF3B1WT) and Panc05.04 (SF3B1K700E) cells. (f) Relative viability of U2AF1S34F/+ or SRSF2P95H/+ knockin cells expressing the indicated synthetic introns. Data represented as mean ± s.d. n = 3 biologically independent experiments. Uncropped gels are available as source data.
Extended Data Fig. 3 Massively parallel screening reveals critical elements governing the function of very short synthetic introns.
(a) As Fig. 3b, but restricted to deletions resulting in an intron of length 100 nt. (b) As Fig. 3c, but for mutations to synMTERFD3i1-100. n = 180 (neither), 90 (any 3’ss), or 30 (5’ss) distinct introns. p computed using a two-sided Wilcoxon rank-sum test. n = 6 replicates (n = 3 independent collections x n = 2 independent sequencing strategies). Multiple testing corrections were not used as only three comparisons were performed. (c) As Fig. 3d, but for mutations to synMTERFD3i1-100. n = 4, 274, and 5 distinct introns for ≤0, 0-7, and ≥7; p computed using a two-sided Wilcoxon rank-sum test. n = 6 replicates (n = 3 independent collections x n = 2 independent sequencing strategies). Multiple testing corrections were not used as only three comparisons were performed. (d) As Fig. 3e, but for mutations to synMTERFD3i1-100. (e) As Fig. 3f, but illustrates synMTERFD3i1-100. (f) As Fig. 3g, but for mutations to synMTERFD3i1-100. Shading indicates error bars, calculated as the geometric standard deviation of the fold changes over replicates. n = 6 replicates (n = 3 independent collections x n = 2 independent sequencing strategies). (g) As Fig. 3h, but for mutations to synMTERFD3i1-100. (h) Box plot illustrating relative fold-changes for introns derived by inserting a very strong 3’ss and key upstream sequence elements (1-4 consensus branchpoints, inserted at positions +25 to +50 relative to the 5’ss, and TTTTTTTTTTTTTTTTTCAG, representing a long polypyrimidine tract immediately followed by a 3’ss) within synMTERFD3i1-100, with 0-8 nt between the last nucleotide of the inserted TTTTTTTTTTTTTTTTTCAG and the canonical 3’ss. p computed using a two-sided Wilcoxon rank-sum test. n = 100 introns per bin. n = 6 replicates (n = 3 independent collections x n = 2 independent sequencing strategies). For all box plots, the middle line, hinges, notches, whiskers, and points indicate the median, 25th/75th percentiles, 95% confidence interval for the median, most extreme data points within 1.5× the interquartile range from the hinge, and outliers.
Extended Data Fig. 4 Branchpoint manipulation and combinatorial 3’ss mutations enhance SF3B1 mutation-dependent splicing.
(a) Diagrams of modifications in each intron relative to synMTERFD3i1-150. Deletions are specified as open intervals. Branchpoints were inserted in between the indicated positions. (b) Relative viability of K562 cells expressing the indicated constructs, measured in the full screen. Relative viability estimated as fold-change in representation of each construct, measured by full-length intron sequencing from genomic DNA, at day 8 for GCV-treated relative to untreated samples. GCV concentration, 100 ug/mL. Vector is hPGK-PuroR-P2A-HSV-TK. Data represented as mean ± s.d; s.d. estimated as sample proportion s.d. over counts. n = 6 replicates (n = 3 independent collections x n = 2 independent sequencing strategies). (c) Relative viability of K562 cells expressing the indicated constructs, measured in cells expressing each construct individually. Viability estimates from these single-construct experiments are concordant with estimates from parallelized screening in (b); note that fold-changes are greater in this experiment because of its longer duration (11 vs. 8 days). Relative viability measured by ATP after 11 days of treatment and normalized to PBS-treated samples. GCV concentration, 100 ug/mL. Vector is hPGK-PuroR-P2A-HSV-TK. Data represented as mean ± s.d. n = 3 biologically independent experiments. (d) RT-PCR demonstrating mutation-dependent excision of the synthetic intron in the experiments from (c). (e) RT-PCR demonstrating mutation-dependent excision of the synMTERFD3i1-150 synthetic intron or control synthetic introns in K562 cells with or without knockin of SF3B1K700E or SF3B1K666N, or MEL202 cells with an endogenous SF3B1R625G mutation. Experiment repeated independently n = 3 times with similar results. (f) RT-PCR of a series of synthetic introns in SF3B1R625G-mutant MEL202 cells. Experiment repeated independently n = 2 times with similar results. (g) RT-PCR of HSV-TK interrupted by synMTERFD3i1-150-v6700 or control synthetic introns in uveal melanoma cell lines that are wild-type or mutant for SF3B1. Experiment repeated independently n = 3 times with similar results. (h) Relative viability of the cells in (g) following treatment with 1 µg/mL GCV. Data represented as mean ± s.d. n = 3 biologically independent experiments. Uncropped gels are available as source data.
Extended Data Fig. 5 Synthetic introns enable mutation-dependent targeting of Sf3b1-mutant hematopoietic cells in vivo.
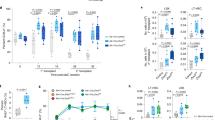
(a) Schematic of xenograft experiments with MOLM-13 cells expressing doxycycline-inducible SF3B1WT or SF3B1K700E. MOLM-13 cells were intravenously injected into sub-lethally irradiated (250 cGy) NOD-scid IL2rgnull (NSG) mice (100,000 cells/mouse). Doxycycline was provided in feed one day before xenograft. (b) Radiance of experiment in (a). Each point represents an individual animal; bars represent means. n = 11 to 15 mice/group. p computed with two-way ANOVA with Tukey’s multiple comparisons test. (c) Schematic of mixed chimeric bone marrow (BM) hematopoietic stem and progenitor experiment. BM cells from CD45.2+ Mx1-cre Sf3b1WT or Sf3b1K700E/WT mice were mixed 3:1 with BM cells from CD45.1+ C57BL/6 wild-type mice. The mixed population was infected with a lentiviral vector encoding GFP-P2A-HSV-TK interrupted by synMTERFD3i1-150. Following two sequential days of infection, cells were treated with GCV in vitro, and GFP+ cell percentages were enumerated by FACS. (d) Schematic of in vivo experiment with primary hematopoietic precursors from Mx1-cre Sf3b1K700E/WT or littermate Sf3b1WT mice. c-Kit+ cells from BM of CD45.2+ Mx1-cre Sf3b1WT or Sf3b1K700E/WT mice were infected with GFP-P2A-HSV-TK interrupted by synMTERFD3i1-150 and intravenously injected into sub-lethally irradiated CD45.1+ wild-type mice. 3 weeks post BM transplantation, CD45.1+ recipient mice underwent pIpC and GCV treatment and were then bled for CD45.2+, CD45.1+, and GFP+ content in peripheral blood and BM. n = 5 (WT) and n = 4 (Sf3b1-mutant) mice per cohort. (e) GFP+ cell percentages amongst CD45.2+ cells in peripheral blood post-pIpC and GCV treatment from cohort in (d). Data represented as mean ± s.d. n = 5 (WT) and 4 (Sf3b1-mutant) mice per cohort. (f) Box plots of GFP+ percentages amongst CD45.2+ cells in peripheral blood at days 0 and 39 post-pIpC and GCV treatment from cohorts in (d). Points depict individual mice. p computed by an unpaired, two-sided t-test. (g) As (f), but for BM at day 39. Points depict individual mice. p computed by an unpaired, two-sided t-test. For all box plots, the middle line, hinges, notches, and whiskers indicate the median, 25th/75th percentiles, and min-max range.
Extended Data Fig. 6 Synthetic introns enable mutation-dependent cancer cell targeting in vivo.
(a) Tumor volumes at day 20 post-xenograft from the experiment illustrated in Fig. 5b (T47D cells expressing doxycycline-inducible SF3B1WT or SF3B1K700E and HSV-TK interrupted by synMTERFD3i1-150 following PBS or GCV treatment). n = 10 tumors/group. Data represented as mean ± s.d. (b) Kaplan-Meier curves from the experiment illustrated in Fig. 5c-d (NSG mice subcutaneously engrafted with MEL285 or MEL202 cells expressing HSV-TK interrupted by synMTERFD3i1-150-v6700 following PBS or GCV treatment). n = 5 mice/group. p computed with log-rank test. (c) Tumor weights from (b) at day 27 (MEL285) or day 30 (MEL202) post-xenograft. n = 10 tumors/group. Data represented as mean ± s.d. p computed with two-sided t-test.
Supplementary information
Supplementary Information
Supplementary Note and Figs. 1 and 2.
Supplementary Table
Supplementary Tables 1–6. Table 1. Differentially spliced events in SF3B1-mutant versus WT cancer samples. Table specifying the splicing events that were most responsive to SF3B1 mutations and so considered for subsequent synthetic intron design (data visualized in Fig. 1b). Each row corresponds to a single isoform of an individual splicing event. Columns are as follows: event, event ID; difference in isoform ratio, mean difference in absolute isoform expression (range from 0 to 1.0) between samples with an SF3B1 mutation and those without any recurrent splicing factor mutations, computed across patient cohorts; standard deviation of difference in isoform ratio; gene ID, Ensembl ID for parent gene; gene name, name of parent gene. All subsequent columns specify the mean difference in isoform ratio between samples with an SF3B1 mutation and those without any recurrent splicing factor mutations within the indicated cohort. See Data availability for descriptions of location of data corresponding to each indicated cohort. Table 2. Characteristics of patient samples studied in Extended Data Fig. 1e. Table describing clinical features of samples from patient cohorts analyzed in Extended Data Fig. 1e by RT–PCR. Table 3. Mini-library composition and results from mini-screen. Table specifying the sequences of each synthetic intron queried in the mini-screen (Fig. 2) and associated fold changes in WT and SF3B1-mutant K562 cells. Each row corresponds to a single fold change measurement for a single synthetic intron. Columns are as follows. id: intron ID; id_verbose: verbose intron ID used in paper (when available); family: intron family; modification: sequence modification applied to derive intron variant; modification_type: type of modification; modification_location: position(s) within intron where modifications were applied; length: intron length in nt; genotype: SF3B1 genotype (WT is SF3B1+/+; K700E is SF3B1K700E/+); fold change: estimated fold change in intron abundance in gDNA at day 6 relative to day 0; s.d., standard deviation of fold change over replicates; sequence: intron sequence. Note that IDs from the mini-library do not correspond to IDs from the full library. Table 4. Sequence modifications represented in full library. Table specifying numerical breakdown of full library by parent synthetic intron and modification type(s) used to create each class of intron variant. See Supplementary Information for detailed descriptions of each modification. Table 5. Full library composition and results from full screen. Table specifying the sequences of each synthetic intron queried in the full screen (Fig. 3) and associated fold changes in WT and SF3B1-mutant K562 cells. Each row corresponds to a single fold change measurement for a single synthetic intron. Columns are as in Supplementary Table 2. Table 6. PCR primers and other oligonucleotides. Table specifying sequences of all oligos used in this study for PCR, cloning and so on.
Source data
Source Data Fig. 1
Uncropped gels.
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Uncropped gels.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Uncropped gels.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Uncropped gels.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Uncropped gels.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Uncropped gels.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
North, K., Benbarche, S., Liu, B. et al. Synthetic introns enable splicing factor mutation-dependent targeting of cancer cells. Nat Biotechnol 40, 1103–1113 (2022). https://doi.org/10.1038/s41587-022-01224-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-022-01224-2
This article is cited by
-
Roles and mechanisms of aberrant alternative splicing in melanoma — implications for targeted therapy and immunotherapy resistance
Cancer Cell International (2024)
-
Variations of intronic branchpoint motif: identification and functional implications in splicing and disease
Communications Biology (2023)
-
Regulation of pre-mRNA splicing: roles in physiology and disease, and therapeutic prospects
Nature Reviews Genetics (2023)
-
Non-canonical functions of spliceosome components in cancer progression
Cell Death & Disease (2023)
-
A trojan horse to target spliceosome mutant cells
Nature Reviews Cancer (2022)